So-SPIM-FCS
By Sruthi Jagannathan, M.Sc. Illustration by Diego Pitta de Araujo, PhD | January 2017
Researchers at the Mechanobiology Institute (MBI), National University of Singapore and the Interdisciplinary Institute for Neuroscience (IINS), France have developed a novel technological approach for observing the complex behavior of proteins inside the nucleus and analyzing their effect on cellular functions. Led by MBI Research Fellow Dr. Anand Pratap Singh and IINS Researcher Dr. Remi Galland along with MBI Principal Investigators Asst. Prof. Timothy Saunders and Assoc. Prof. Virgile Viasnoff, the study combined principles of high resolution imaging and analytical techniques such as single objective selective plane illumination microscopy (So-SPIM) and fluorescence correlation spectroscopy (FCS) to measure protein dynamics within the three-dimensional nuclear space.

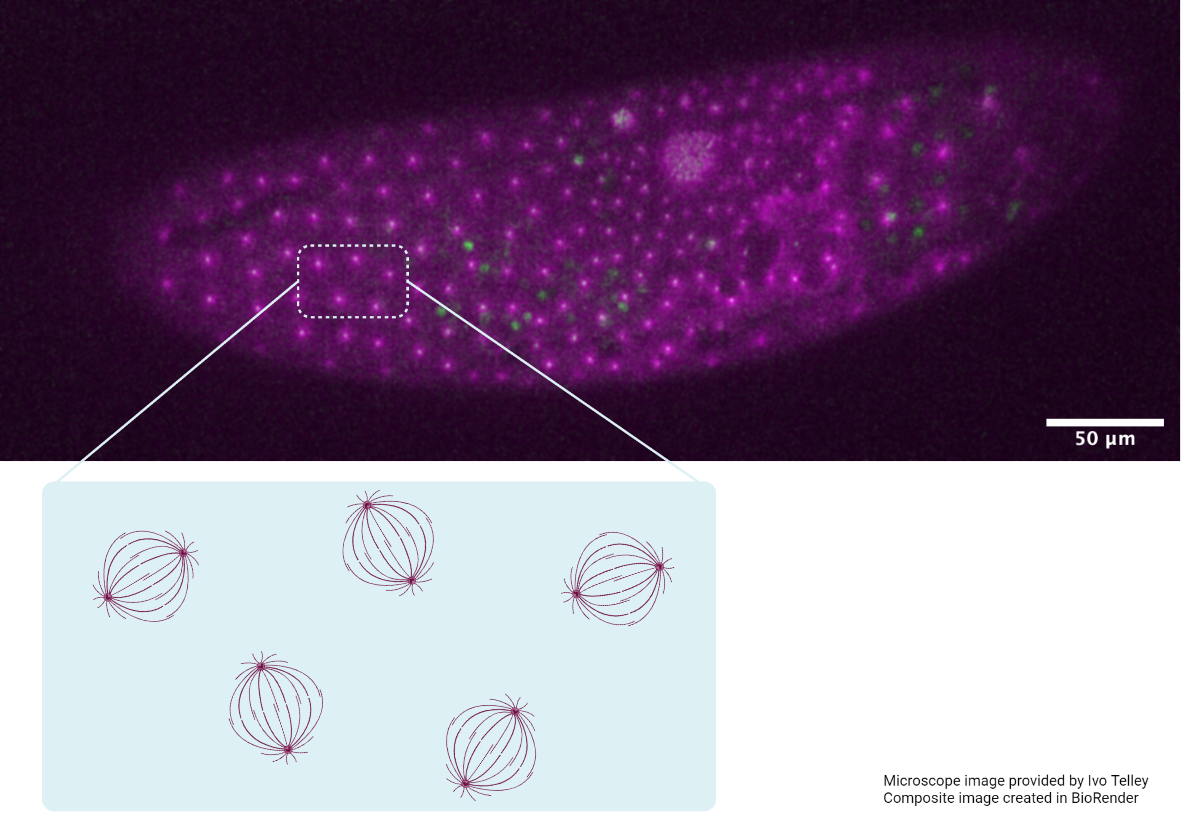
A schematic representing the visualization of nuclear protein dynamics using the so-SPIM-FCS technique.
A novel tool for imaging nuclear protein dynamics
The nucleus is the information processing center of a eukaryotic cell. It contains various sub-nuclear compartments, each of them housing specific DNA regions and proteins. The diverse functions of the nucleus are facilitated by the sharing of protein machineries between nuclear compartments. A vast number of nuclear proteins roam the nucleus, rapidly associating and dissociating with other proteins or DNA segments. This enables the nucleus to process different signal types, such as chemical, mechanical, and electrical signals, and in response, turn specific genes on or off.
Tracking protein movement in time and space
Observing the movement of proteins inside the 3D space of the nucleus would reveal valuable information on their specific biological roles. Despite the advent of high power imaging techniques that allow researchers to visualize the dynamics of fluorescently labeled single proteins in living cells, their use has been limited due to their phototoxic effects on cells. Phototoxicity is induced by prolonged exposure to light, whereupon fluorescent molecules, in their excited states, tend to react with molecular oxygen and form free radicals that can damage subcellular structures.
The data from the so-SPIM-FCS measurements clearly highlights how protein activities constantly vary inside the nucleus, at various points in time and space.
In order to get past this problem, researchers at the Mechanobiology Institute, Singapore and the Interdisciplinary Institute for Neuroscience, France combined a microscopy method that they had recently developed, known as so-SPIM, with FCS, an analytical technique for measuring protein dynamics.
What is so-SPIM and FCS?
So-SPIM allows the sample (a single cell in this case) to be illuminated along a narrow focal plane and images are captured by an objective lens placed perpendicular to the sample. The sample is then moved through the light beam to expose different parts or sections for visualization at any given time. Since this technique restricts the light beam to a narrow section of the sample, so-SPIM minimizes phototoxicity.
FCS measures the brightness of light emitted by proteins tagged with fluorescent dyes at different points in time. Any variations in brightness over the observation period is linked to the movement of proteins in time and space.
How can these methods overcome phototoxicity?
By combining the two techniques, the researchers were able to measure the brightness of light emitted by fluorescently-labeled proteins at multiple spatial locations within the nucleus. Any fluctuations in brightness were inferred as changes in protein concentrations within the nuclear area under observation. Using this data, the researchers were able to identify the movement of proteins within the 3D space of the nucleus over a set period of time. Specifically, the proteins moved from regions where they were highly concentrated, to regions of lower concentration. When movement was slow, the researchers could tell that the protein was interacting either with other proteins, or with strands of DNA or RNA. In these cases the protein would be catalyzing gene expression or protein synthesis.
Applying the technique
The researchers used the soSPIM-FCS setup to observe the dynamics of two proteins inside the nucleus, namely RNA Polymerase II and YAP. RNA Polymerase II (Pol II) is a transcription factor- it is responsible for decoding information on DNA into intermediary RNA molecules, which is then translated into proteins. YAP, on the other hand, is a transcriptional coactivator. It binds to transcription factors such as Pol II to facilitate their transcriptional activity.
Interestingly, both Pol II and YAP demonstrated mixed patterns of movement within the nucleus. When they were not interacting with DNA or other proteins, they moved rapidly. However, when they bound to nuclear components at defined spatial locations, their movement slowed down. Using so-SPIM-FCS, the scientists were able to record more than 3000 measurements for each protein, which allowed them to precisely predict protein dynamics across the 3D space of the nucleus.
The capability of so-SPIM-FCS to produce clear images of subcellular structures along multiple planes, whilst minimizing phototoxic effects on cells under study, makes it an ideal technique to track single protein dynamics inside the nucleus. This is displayed here, with the so-SPIM-FCS technique being used for the first time to unravel the dynamics of proteins such as YAP inside the nucleus. The data from the so-SPIM-FCS measurements clearly highlights how protein activities constantly vary inside the nucleus, at various points in time and space. This is critical to understand how dynamic protein behavior influences nuclear organization, and as a result, controls cellular behavior at a global scale.
This study is published in Biophysical Journal [Singh PA, et al. 3D protein dynamics in the cell nucleus].