Finding order through repulsion
A collaborative study demonstrates how mechanical repulsive forces lead to precise arrangement of nuclei during development
While it is typically thought of that a single cell contains one nucleus, there are many occasions in biology where a single cell can contain several nuclei. This is called a syncytium (from the ancient Greek of syn meaning together, and cyte meaning cell) and can occur when the nucleus divides without cell division, or when several cells fuse together to form a cell mass. The classic example in animals of the latter is found in muscle. Skeletal muscle fibres form by fusion of many individual muscle cells, and in the heart, the syncytium of cardiac muscle connects cells together so that they rapidly beat in synchrony.
Syncytia also commonly occur during development. During development of an insect embryo syncytia form with hundreds of nuclei, and when a specific nuclear density is reached the process of cellularisation occurs to generate the first tissues. A unique feature of this is that distribution and location of nuclei in embryos is highly uniform – each nucleus is spaced apart precisely according to a set distance, much like how eggs are packaged in a carton. This lattice-like arrangement and nuclear spacing is important for gene patterning and subsequent cell fate determination. However, the mechanism by which nuclei organise into these lattice-like arrangement remains unknown. It is thought that arrangement of cytoskeletal components such as actin filaments and microtubules can serve to precisely segregate nuclei. In particular, much attention has been focused on the mitotic spindle, a spherical assembly of microtubules that organises and separates duplicated chromosomes during cell division. Kinetochore microtubules attach to the chromosomes, and astral microtubules radiate out from the poles of the spindle like a star to form asters.
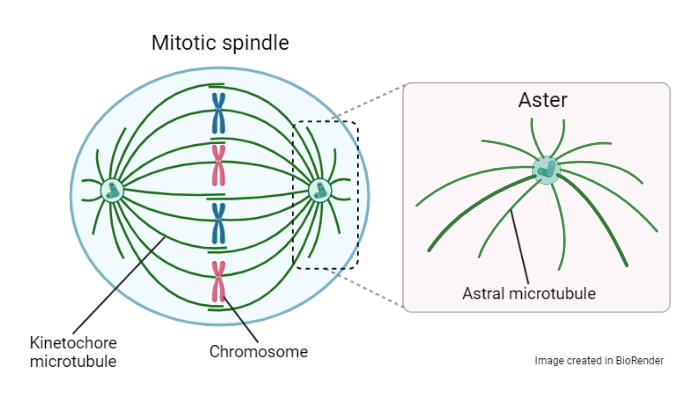
Schematic of the mitotic spindle, showing the asters at the poles of the spindle.
Determining how mechanics control precise nuclear arrangement
To try and discover the mechanisms that control the regular distancing of nuclei, Ivo Telley from Instituto Gulbenkian de Ciencia, Portugal and Timothy Saunders, Mechanobiology Institute, National University of Singapore and University of Warwick, UK, co-investigators of a Human Frontier Science Program (HFSP) Young Investigator team set up an international collaboration. By bringing together complementary scientific expertise, their joint research team utilised a multiscale approach to visualise cellular and tissue-level processes in development, supported by theoretical modelling.
Live imaging of Drosophila fruit fly embryos allowed the researchers to track nuclei and spindle position and measure the distance between neighbouring nuclei. This confirmed that a constant spacing between nuclei was maintained, even when spindles increased in size. Given that nuclei never collide, this suggested to the research team that some short distance repulsion between nuclei or the spindle must be responsible for this organisation. By analysing the position of the nuclei and spindles, they discovered that nuclei self-organised over short distances to form a chain. Specifically, the spindles aligned so that the asters were orientated perpendicular to the direction between neighbouring nuclei.
To gain deeper insight, the researchers were able to carefully extract the cellular material of an embryo by micropipette, thereby removing any potential influence of mechanical force from the surrounding cell cortex, or compartmentalisation by actin filaments. The nuclei were still able to self-organise and, even more remarkably, when the nuclei were removed the spindles continued to organise in a defined lattice-like arrangement. This provided strong evidence that the asters are responsible for controlling this self-organisation. After analysing their experimental data, the research team developed a theoretical model where short-range repulsive interactions between asters can lead to the precise organisation of nuclei. In this model, the asters at the poles of the spindle repel the asters of neighbouring spindles over a short distance, in a similar way to magnetic repulsion. These repulsive interactions move the spindles around until a positional balance is achieved, resulting in a precise spacing of nuclei in a lattice.
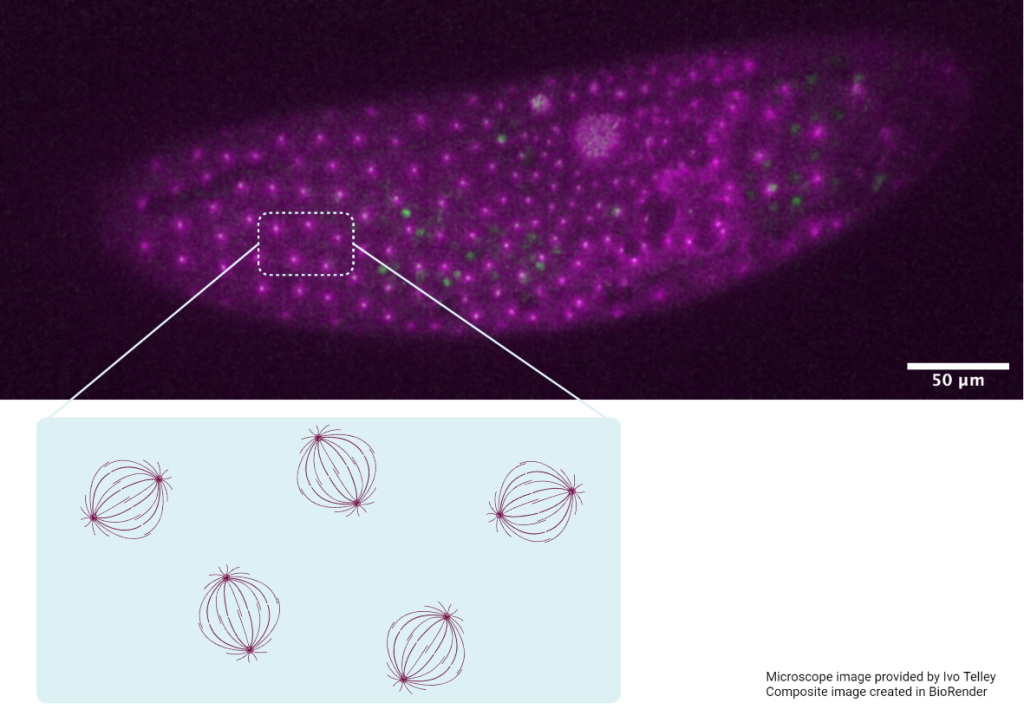
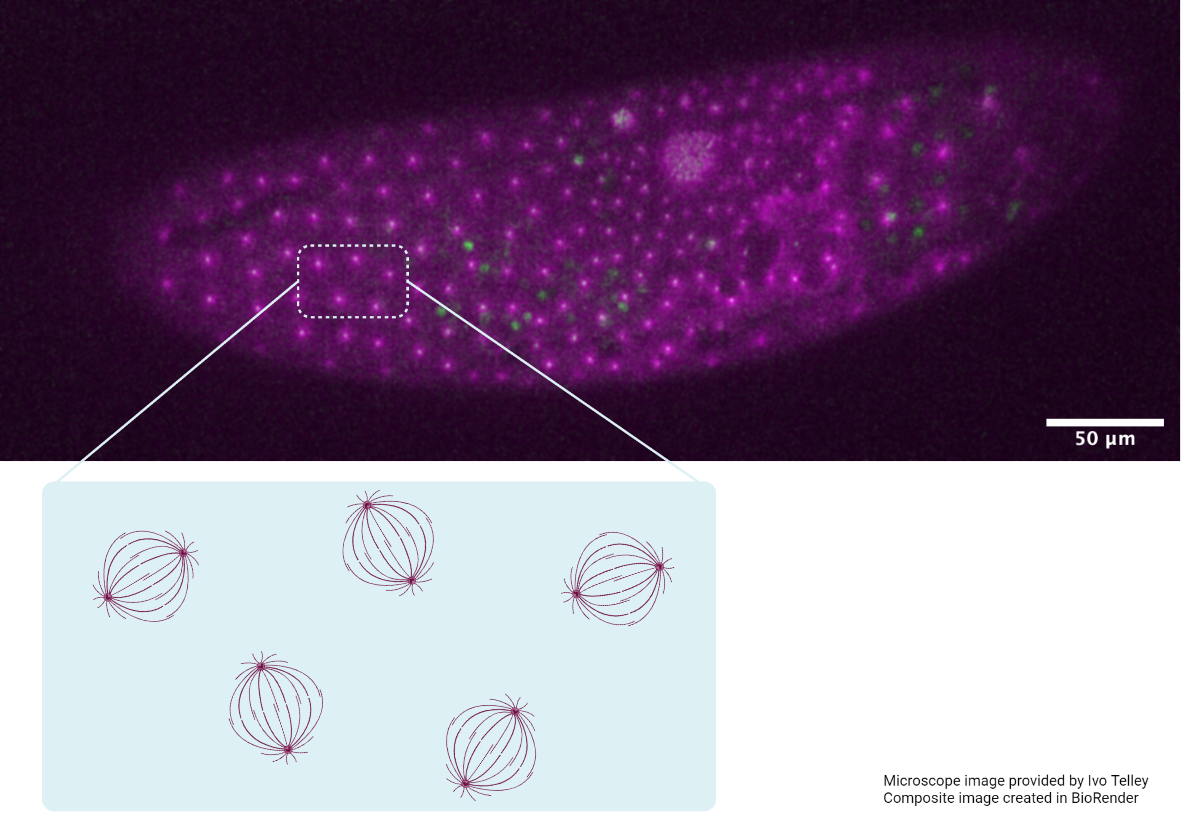
The image shows an embryo from a mutant fruit fly, in which the microtubule cytoskeleton (magenta) and centrosomes (green) were stained. In this mutant, the division of nuclei (chromosomes) is inhibited and an endless growing nucleus forms (middle). Although mitosis does not occur, the spindle asters continue to organise in a lattice-like arrangement, highlighted in the schematic. This provides strong evidence that the asters alone are responsible for regular positioning of spindles and nuclei in wild type flies. The spacing is defined by a repulsion force between neighbouring asters.
In order to test their theoretical model, the research team carried out an experiment where a single nucleus was rapidly removed by laser ablation. The model accurately predicted the observed rearrangement of the remaining nuclei, validating the idea that mechanical repulsion is the driver of nuclei positioning. Importantly, this short-range mechanical repulsion can have far reaching effects in organising nuclei across the embryo. This robustness also allows for dynamic adaptation and maintenance of positioning in case of a sudden increase or decrease in nuclei number. Development is a complicated process of cell division, positioning, and cell differentiation into specialised tissues and organs. The repulsion-driven mechanism described in this paper demonstrate how biology utilises mechanical forces as a safeguard for ensuring correct biological processes.