Saradha VENKATACHALAPATHY
PhD Student, Mechanobiology Institute, National University of Singapore
E0018317@u.nus.edu
Level 10 T-Lab
National University of Singapore
5A Engineering Drive 1
Singapore 117411
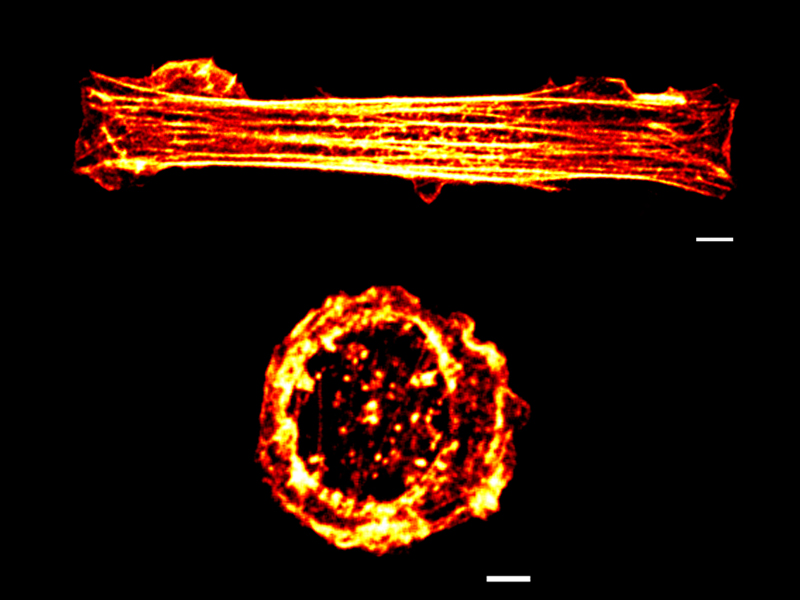
Physically rewiring the genome
Researchers turn mature cells into stem cells using mechanical cues alone
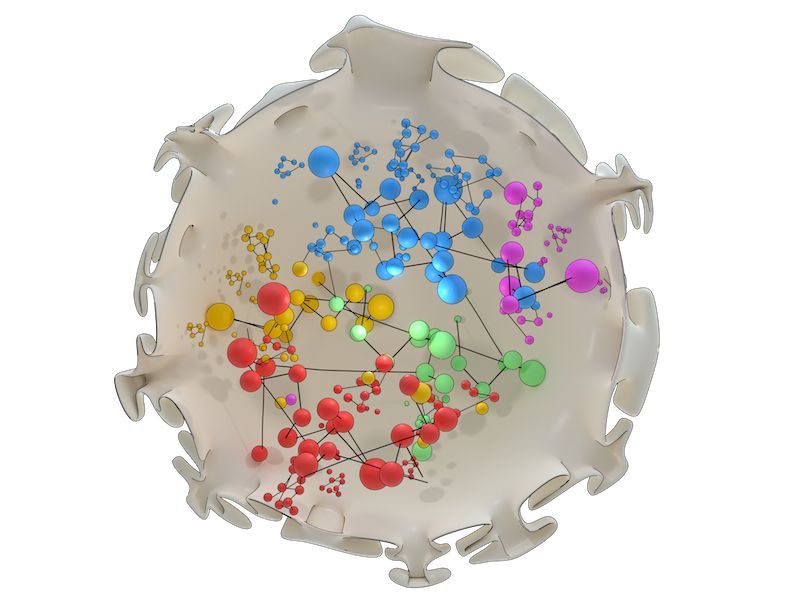
Determining genomic architecture
Network analysis reveals 3D chromosome organization
Signaling in 3D
MBI Scientists reveal a spatial dimension to cell signaling
Saradha Venkatachalapathy
PhD Student
Recent Publications
- Venkatachalapathy S, Sreekumar D, Ratna P, and Shivashankar GV. Actomyosin contractility as a mechanical checkpoint for cell state transitions. Sci Rep 2022; 12(1):16063. [PMID: 36163393]
- Venkatachalapathy S, Jokhun DS, Andhari M, and Shivashankar GV. Single cell imaging-based chromatin biomarkers for tumor progression. Sci Rep 2021; 11(1):23041. [PMID: 34845273]
- Yang KD, Belyaeva A, Venkatachalapathy S, Damodaran K, Katcoff A, Radhakrishnan A, Shivashankar GV, and Uhler C. Multi-domain translation between single-cell imaging and sequencing data using autoencoders. Nat Commun 2021; 12(1):31. [PMID: 33397893]
- Yang KD, Damodaran K, Venkatachalapathy S, Soylemezoglu AC, Shivashankar GV, and Uhler C. Predicting cell lineages using autoencoders and optimal transport. PLoS Comput. Biol. 2020; 16(4):e1007828. [PMID: 32343706]
- Venkatachalapathy S, Jokhun DS, and Shivashankar GV. Multivariate analysis reveals activation-primed fibroblast geometric states in engineered 3D tumor microenvironments. Mol. Biol. Cell 2020;:mbcE19080420. [PMID: 32023167]
- Damodaran K, Venkatachalapathy S, Alisafaei F, Radhakrishnan AV, Sharma Jokhun D, Shenoy VB, and Shivashankar GV. Compressive force induces reversible chromatin condensation and cell geometry dependent transcriptional response. Mol. Biol. Cell 2018;:mbcE18040256. [PMID: 30256731]
- Roy B, Venkatachalapathy S, Ratna P, Wang Y, Jokhun DS, Nagarajan M, and Shivashankar GV. Laterally confined growth of cells induces nuclear reprogramming in the absence of exogenous biochemical factors. Proc. Natl. Acad. Sci. U.S.A. 2018;. [PMID: 29735717]
- Belyaeva A, Venkatachalapathy S, Nagarajan M, Shivashankar GV, and Uhler C. Network analysis identifies chromosome intermingling regions as regulatory hotspots for transcription. Proc. Natl. Acad. Sci. U.S.A. 2017;. [PMID: 29229825]
- Radhakrishnan AV, Jokhun DS, Venkatachalapathy S, and Shivashankar GV. Nuclear Positioning and Its Translational Dynamics Are Regulated by Cell Geometry. Biophys. J. 2017; 112(9):1920-1928. [PMID: 28494962]
- Mitra A, Venkatachalapathy S, Ratna P, Wang Y, Jokhun DS, and Shivashankar GV. Cell geometry dictates TNFα-induced genome response. Proc. Natl. Acad. Sci. U.S.A. 2017;. [PMID: 28461498]