Myosin motors show their muscle
How cells generate high forces for testing the softness of their environment
Written by Andrew MS Wong, PhD | Illustration by Diego Pitta de Araujo, PhD | June 2019
Researchers from the Mechanobiology Institute, National University of Singapore discover that the myosin motors inside cells generate forces significantly higher than previously thought. Using theoretical modelling, they explain how these large forces arise and enable individual myosins to work as a collective motor that powers the machinery behind rigidity sensing, a key process for cell growth and movement, which is often hijacked during cancer cell invasion. This study is published in Nature Physics.

Artistic illustration of myosin motors pulling on actin filament ‘chains’ attached to integrin ‘anchors’ that pinch the environment to test its rigidity.
Powering super-strong pinching
One of the major factors which determines how fast a cell can move is the rigidity of the surface it is on. This information can also regulate cell growth, death, and fate choices. Cells sense rigidity by ‘pinching’ their environment, much like you would do with your index finger and thumb when you want to know if something is soft or rough. This pinching occurs at adhesion sites, protein complexes where the cell attaches to the surface. Within these adhesions are integrin proteins, which span across the membrane and connect to the external surface. These integrins are connected by the linker protein a-actinin to the cell’s internal scaffold of actin filaments and myosin motor proteins. Energy derived from the breakdown, or hydrolysis of the molecule adenosine triphosphoate (ATP) powers the stepwise movement of myosin motor heads along actin filaments, and this generates contractile force that powers the pinching activity of integrins, thereby enabling rigidity sensing.
The force generation of the actomyosin contraction process has been studied extensively. Single-molecule experiments that measure pulling force have revealed that a single myosin motor head can generate a force ranging from 1.3-4pN. However, as these in vitro experiments were performed by reconstituting the protein complex outside of the cell, it remained unclear what the force generation process is like inside a living cell.
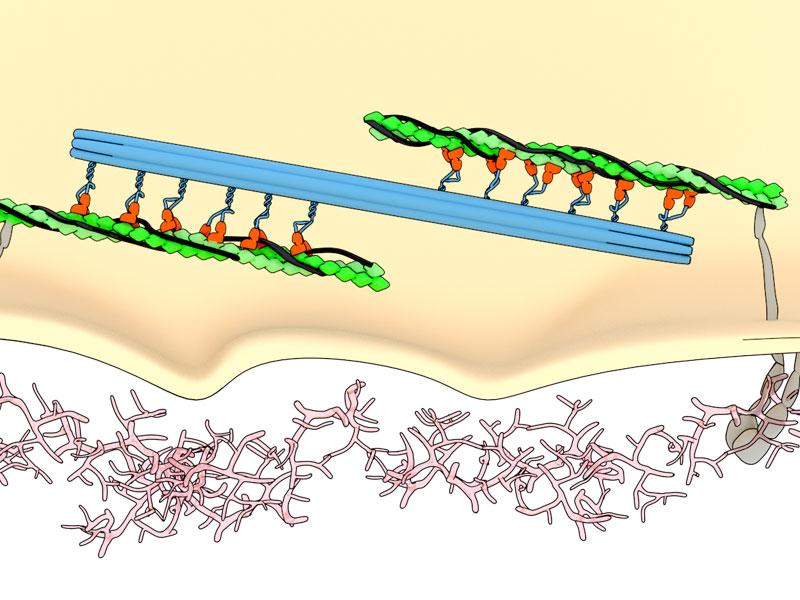
Illustration of rigidity sensing. Contractile force generated by movement of myosin heads (orange; attached to the myosin thick filament (blue)) on actin filaments (green) powers the pinching activity of the transmembrane protein integrin (grey).
Understanding this process required a joint experimental and theoretical study, led by MBI Professors Michael Sheetz and Jacques Prost, in collaboration with colleagues from Columbia University, USA. In order to measure force generation in connective tissue fibroblast cells, the research team created a surface of pillars made from an elastic material known as PDMS. When PDMS is exposed to ultraviolet light it stiffens, and this property was harnessed by the researchers to design a chessboard arrangement of alternating areas of soft and stiff pillars, which had a 20-fold difference in stiffness (for comparison, a diamond is roughly 20-times stiffer than an aluminium can). This clever chessboard arrangement eliminates experimental variability, as a single cell is exposed to both soft and stiff surfaces at the same time. After growing cells on this chessboard surface, the scientists were able to measure the displacement of pillars by the contractile unit. Interestingly, cells displaced soft and stiff pillars by the same distance of 60nm, in 2.5nm steps. Careful examination of the myosin motor revealed that similar numbers of myosin heads were active when contracting on both soft and stiff pillars. By dividing the total force exerted on the pillars by the number of motors, they estimated that each myosin head could generate a force up to 60 pN.
MBI researchers discover how and why myosin motors generate huge forces in the living cell.
This maximal output was significantly higher than the predicted force ranges from experiments in vitro, leading the researchers to investigate the possible reasons behind this >10-fold difference. The conventional understanding of myosin head movement is that the energy derived from the hydrolysis of a single ATP molecule provides the energy needed to perform a myosin step contraction. However, within this framework, the corresponding maximal force was still significantly lower than what is measured within cells. Since larger force generation requires larger energy consumption, the team suggested that more than a single ATP hydrolysis event should be needed to make a step. A possible scenario is that most ATP hydrolysis events would fail to result in a step, but due to thermal fluctuations a fraction of ATP hydrolysis events succeed in performing a myosin step. Under this scenario, the contractile unit still respects the fundamental laws of thermodynamics, as the total energy consumed in ATP hydrolysis, including unsuccessful ATP hydrolysis events, remains lower than the mechanical energy released during a step.
The next question
But this led to the next question: how could all myosin motors synchronize with each other to generate such forces? To answer this question, the team revisited a classical molecular motor model [J. Prost & F. Julicher, 1995]. MBI Senior Research Fellow Dr. Jean-Francois Rupprecht considered the effect of large binding & unbinding energies using this model. Using this framework, the team was then able to successfully simulate and validate their experimental findings, and also revealed that the pinching dynamics enable individual myosins to work as a collective motor.
By measuring forces at the single-molecule level in living cells, this study extends our understanding of how cells sense the rigidity of their environment. Since rigidity sensing is perturbed in many cancer cells, allowing them to grow and move abnormally rapidly on soft surfaces, the findings from this study could pave the way for a better understanding and improvement for current therapeutic treatments that revolve around inhibition of tropomyosin, which surrounds actin and regulates myosin binding, potentially increasing the energy barrier to actomyosin force generation.









