Digitalising Tissue Biology: How AI is Helping Scientists Map 3D Cellular Landscapes
DeepStar3D, a robust AI model trained on a huge dataset of simulated images accurately detects cell nuclei, maps cell boundaries, and outlines entire organoids before sequentially stacking layers of segmented data.
Researchers combine DeepStar3D algorithm and 3DCellScope interface to develop an experimental tool for real-time interaction with cell data. The method promises to support research in ares of tissue regeneration, drug testing, and even space biology. This study was recently described in a Nature Methods paper. https://doi.org/10.1038/s41592-025-02685-4
An innovative digitalised organoid approach that can map individual cells within an organoid has been developed by a team of researchers from the Mechanobiology Institute (MBI), National University of Singapore, in collaboration with institutions from France and USA. Led by Hui Ting Ong and Dr Esra Karatas from MBI’s microscopy core facility, and guided by facility manager, Asst Prof Anne Beghin, the team’s novel approach integrates an AI-based algorithm with an easy-to-use 3DCellScope interface. Their work was recently described in a Nature Methods paper [https://doi.org/10.1038/s41592-025-02685-4].
In the human body, cells do not exist in isolation or float around randomly – they organize into definitive patterns and structures that form tissues and organs, whose coordinated functions are crucial for a normal physiological state. To achieve and sustain this structural arrangement, cells sense the mechanical properties of their surroundings and constantly adjust their shape, position and interactions accordingly. However, in responsed to aberrant mechanical signals such as during ageing or disease conditions, cell properties and behaviour are disrupted, leading to loss of tissue equilibrium. Understanding how individual cells respond to mechanical changes is therefore critical to gaining insights into tissue functions during healthy and diseased states.
Recent advancements in the development of lab-grown mini-organ systems – such as organ-on-chips or organoids – have enabled deeper investigations into tissue functions. However, detailed study of the morphology and functions of individual cells within the organoid has been limited. The digitalised organoids approach described by the research team addresses this gap. By using a powerful AI algorithm, it segments 3D microscopy images at multiple scales – from entire organoids down to single-cell and nuclear levels. A single scan with this high-resolution segmentation tool enables precise mapping of every single cell in the organoid, reveal hundreds of biological features and tracks changes in their shapes and arrangement under different conditions.
At the core of this approach is DeepStar3D, a robust AI model trained on a huge dataset of simulated images that closely resemble those obtained in the lab. It accurately detects cell nuclei, maps cell boundaries, and outlines entire organoids before sequentially stacking layers of segmented data. The layered 3D images are then analysed using advanced data mining tools, while the user-friendly 3DCellScope interface facilitates visualisation and analysis – offering crucial insights into how cells organize, behave and respond to mechanical cues.
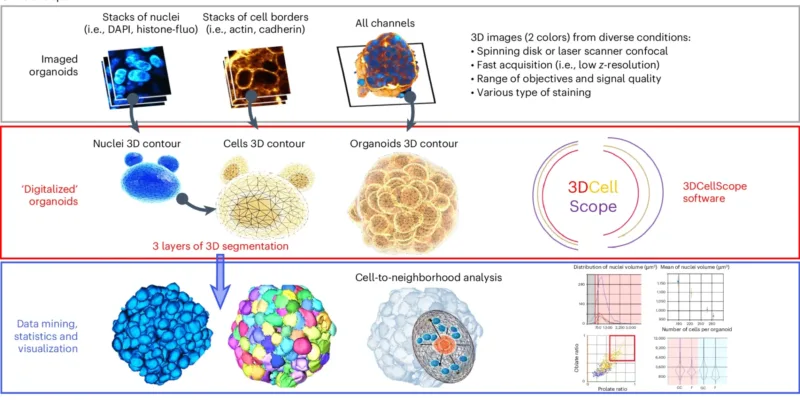
Schematic depicting the pipeline. Top, Organoids were imaged using 3D fluorescence microscopy. Middle, The complete 3D digitalization of organoids consists of three layers of segmentation. Bottom, The resulting data are understandable through data mining. Image from: https://doi.org/10.1038/s41592-025-02685-4
To test the ability of 3DCellScope in detecting cell changes, the researchers simulated physiological conditions involving mechanical stress. Under compressive forces such as osmotic stress, cells became more rounded, and their nuclei shifted towards the centre of the organoid. Cells arrangement also seemed to be impacted by the topology of spaces in which they grew: in tighter, cup-shaped spaces they grew in an organized arrangement, but in flatter spaces and cancer spheroids the cells exhibited a more scattered or random arrangement. Furthermore, when subjected to cyclic mechanical stress introduced by transporting breast cancer spheroids on parabolic flights that mimic weightlessness, cells at the periphery shrank in volume, while those at the core remained relatively unaffected.
Beyond the user-friendly interface, 3DCellScope other advantages over existing tools. It is compatible with standard laboratory conditions , requires no complex AI training or image staining methods. Its flexibility allows integration with open-source AI algorithms, fostering innovation and collaboration with the broader research community. Furthermore, it accommodates real-world image variability, delivering accurate results across microscopy images that differ in resolution, staining methods, or cell types.
Owing to its simple set up and powerful performance, the digitalised organoid approach, with its 3DCellScope interface, is poised to become a reliable and standard experimental tool for real-time interaction with cell data. By revealing subtle changes in cell shapes, patterns, and interactions in 3D, it promises to enhance our understanding of dynamic and complex physiological and pathological processes, and to support research in areas of tissue regeneration, drug testing, and even space biology.