Controlling how bacteria respond to stress
Bacterial self-protection by self-acidification
Written by Andrew MS Wong, PhD, Illustration by Melanie Lee, PhD | March 2018
A research team from the Mechanobiology Institute, National University of Singapore have discovered a new mechanism that controls how Salmonella and Escherichia coli acidify internally in response to acidic and osmotic stress. Using single cell analysis, they also revealed that this self-acidification is maintained for a prolonged period of time, in contrast to the expected wisdom that bacteria rapidly acidify and recover their pH. This study was published in Nature Communications.

Both Salmonella and E. coli bacteria self-acidify internally following acid or osmotic stress. This is controlled by the EnvZ-OmpR two component regulatory system. Membrane bound EnvZ sensor protein (purple) physically interacts with the OmpR response regulator protein (light blue). OmpR then assembles with itself to form a dimer, which binds to DNA (yellow helix) to control gene expression of pathways that lead to cytoplasmic acidification.
Unmasking the bacterial stress response
Bacterial cells are subjected to a number of stresses throughout their life as they attempt to grow and reproduce. This can include dramatic changes in osmolarity (the concentration of a solution) and pH (acid stress), which they encounter as they move through different environments during the infection process, or as the host organism mounts an immune response. In order to survive, bacteria have evolved a number of strategies to protect themselves. In 2015, research from the lab of Prof. Linda Kenney at MBI revealed that Salmonella Typhimurium self-acidified internally in response to external acid stress, and that this acidification serves as a trigger for the bacteria to activate virulence genes that promote survival and reproduction.
By measuring pH levels in individual bacteria, MBI researchers reveal how bacteria self-acidify and remain acidified in response to stress, overturning the conventional understanding that bacteria restore their pH to normal levels after acidification.
This adaptability relies on a bacterial two component regulatory (TCR) system, which in this case is comprised of the sensor protein EnvZ and the response regulator protein OmpR. In a classic TCR system, the sensor protein detects an environmental change (e.g. pH level, osmolarity, temperature), and becomes activated by phosphorylation. This phosphate group is then transferred to activate the response regulator protein, which then binds to DNA and alters gene expression so that the bacteria can adapt to the change.
Following on from their previous study, the research team wanted to find out whether this response was unique to Salmonella or shared by other bacteria, and to determine the mechanism by which bacteria sense and respond to stressors. In order to answer these questions, they used a fluorescent biosensor to measure pH levels inside individual Salmonella and E. coli bacteria in response to acid or osmotic shock.
Bacterial self-acidification following acidic and osmotic stress
They discovered that Salmonella and E. coli respond to both acidic and osmotic stress. Both stresses caused the bacterial interior to acidify, but to differing degrees and via different pathways. The internal pH of both bacteria became around 10 times more acidic following acid stress, and a similar but less dramatic response occurred following osmotic stress. Interestingly, single cell analysis revealed that bacteria remain acidified after stress, in stark contrast to more than 30 years of research which suggested that the internal cytoplasmic pH of bacteria rapidly acidifies but then recovers to pre-stress levels. This self-acidification is controlled by OmpR, as removal of the response regulator protein prevented both bacteria from acidifying.
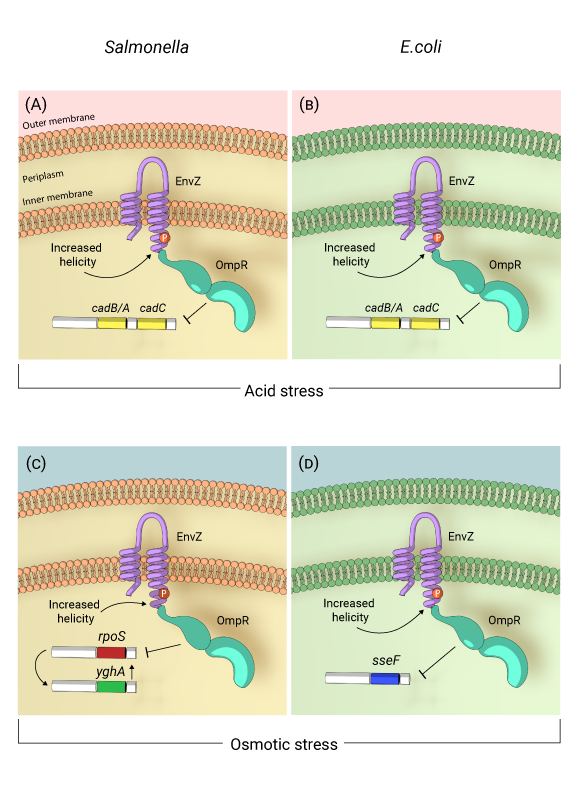
Self-acidification of Salmonella and E.coli in response to acid and osmotic stress: In response to acidic environment resulting from either acid or osmotic stress, EnvZ, the membrane bound sensor kinase is activated by increased helicity of a region surrounding the phosphorylated histidine (P). Activated EnvZ physically interacts with OmpR to promote OmpR dimerization and DNA binding. In both Salmonella (A) and E.coli (B), OmpR represses the cadC/BA operon in response to acid stress to maintain an acidic cytoplasm. In response to osmotic stress in Salmonella, OmpR represses rpoS, which leads to up-regulation of the oxidoreductase yghA, leading to proton production and cytoplasmic acidification (C). In E.coli, OmpR represses speF which codes for ornithine decarboxylase to maintain an acidic cytoplasm under osmotic stress (D).
With the knowledge that OmpR controlled both the acidic and osmotic stress response in bacteria, the researchers subsequently investigated how OmpR exerts this control. They discovered that the initial acid stress activated OmpR, which was then able to prevent decarboxylase proteins from mopping up acidic protons, leading to cytoplasmic acidification. Following osmotic stress, a similar pathway was observed in E. coli. However, in Salmonella, the activated OmpR switches on a pathway of proteins that produce protons. Whether it was by repressing carboxylase proteins or by activating proton production, OmpR acts as a switch regulated by the pH threshold level that acidifies the bacteria in response to acid or osmotic stress.
Activating two-component regulatory systems without phosphorylation
Remarkably, a mutant OmpR that could not be activated by phosphorylation was still able to acidify the bacterial cytoplasm, indicating that it was functioning by a different mechanism than the classic TCR system. While the presence of the EnvZ sensor was required, the scientists discovered that un-phosphorylated OmpR can activate, assemble with itself to form a dimer, bind to DNA and regulate gene expression. This challenges the long-standing notion of how TCRs operate, suggesting that some systems can work without the need for activation by phosphorylation.
One of the keys to these new findings was the ability to measure the response in individual bacteria. Previous experimental studies on bacterial stress response have relied on measurements taken from populations of bacteria. However, in the same way that recording the average age or height of a population masks those characteristics when it comes to individual people, it became clear from this study that single cell analysis was necessary to reveal the full story of how bacterial pH is regulated following stress.
This study revealed a new mechanism for activation of OmpR in response to acid and osmotic stress that does not rely on phosphorylation. With this landmark finding, a completely new avenue of investigation has been opened up for understanding how bacterial TCR systems work. Furthermore, this activation of OmpR functions as a pH-sensitive switch that drives the bacterial response to acidic or osmotic stress. Both stresses result in acidification of the bacterial cytoplasm, which is essential for stimulation of virulence genes, raising the possibility for anti-bacterial therapies based on OmpR regulation.