Linda J KENNEY
Alumni, Mechanobiology Institute, National University of Singapore
H-NS Proteins as Gene Silencers
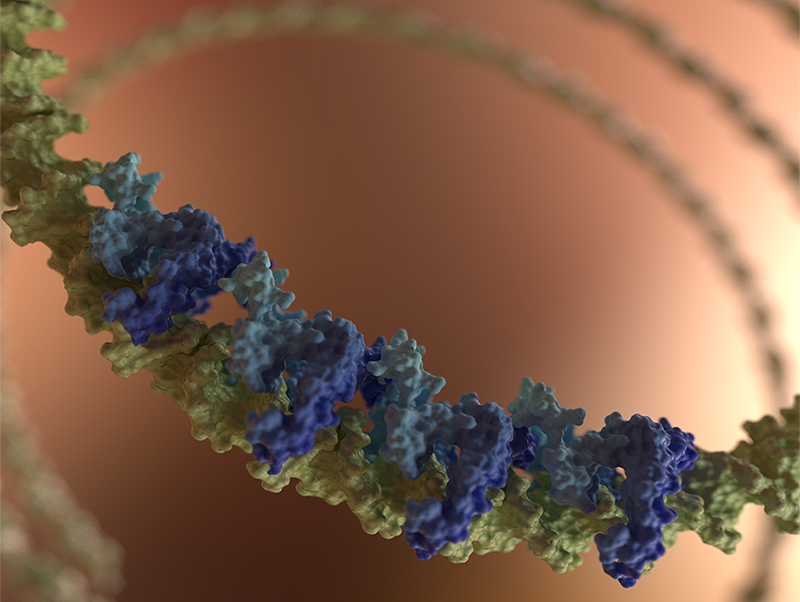
A gene-silencing nucleoprotein filament formed by H-NS family proteins
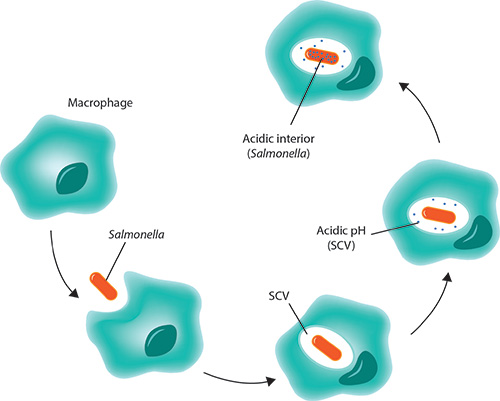
Surviving Immune Attack
How does Salmonella thrive in the acidic macrophage?
Linda J Kenney
Alumni
Research Areas
Signal transduction in bacteria; Bacterial Pathogenesis; Mechanotransduction and osmotic signaling in E. coli; Mechanisms of anti-silencing of virulence genes in Salmonella
Research Interests
Our laboratory is interested in signal transduction and the regulation of gene expression in prokaryotes. In particular, we are studying the two-component regulatory system EnvZ/OmpR that regulates the expression of outer membrane proteins as well as many other genes. Our present work focuses on how OmpR activates genes required for systemic infection (located on Salmonella pathogenicity island 2) in Salmonella enterica.
The Kenney Lab’s research was recently featured in an article exploring the bacterial molecular switch between virulence or dormancy, Salmonella Lifestyle Choices. Members of her lab have discovered that the bacterial protein SsrB is the molecular switch for determining whether Salmonella infections become acute and virulent, or remain in a dormant carrier state.
The study is published in eLife (Desai et al., The horizontally-acquired response regulator SsrB drives a Salmonella lifestyle switch by relieving biofilm silencing, February 2, 2016, eLife 2016; 5: e10747, doi: 10.7554/eLife.10747). Read full article.
Biography
Dr Kenney is a Professor of Microbiology at the University of Illinois-Chicago. Her laboratory studies two-component systems in bacteria that control gene expression at a single cell and nanometer level.

Professor Linda J Kenney and Professor Michael Sheetz interviewed by the Washington Post at the April 2017 March for Science.
Education
PhD University of Pennsylvania
Recent Publications
- Fernandez M, Yamanaka Y, Zangoui P, White MA, and Kenney LJ. The sulfur assimilation pathway mitigates redox stress from acidic pH in Salmonella Typhi H58. mBio 2025;:e0046725. [PMID: 40422406]
- Shetty D, and Kenney LJ. A pH-sensitive switch activates virulence in Salmonella. Elife 2023; 12. [PMID: 37706506]
- Oh D, Liu X, Sheetz MP, and Kenney LJ. Small, Dynamic Clusters of Tir-Intimin Seed Actin Polymerization. Small 2023;:e2302580. [PMID: 37649226]
- Mon KKZ, Si Z, Chan-Park MB, and Kenney LJ. Polyimidazolium Protects against an Invasive Clinical Isolate of Salmonella Typhimurium. Antimicrob Agents Chemother 2022;:e0059722. [PMID: 36094258]
- Jacob H, Geng H, Shetty D, Halow N, Kenney LJ, and Nakano MM. Distinct Interaction Mechanism of RNAP and ResD and Distal Subsites for Transcription Activation of Nitrite Reductase in Bacillus subtilisψ. J Bacteriol 2021;:JB0043221. [PMID: 34898263]
- Singh MK, Zangoui P, Yamanaka Y, and Kenney LJ. Genetic code expansion enables visualization of Salmonella type three secretion system components and secreted effectors. Elife 2021; 10. [PMID: 34061032]
- Kenney LJ, and Anand GS. EnvZ/OmpR Two-Component Signaling: An Archetype System That Can Function Noncanonically. EcoSal Plus 2020; 9(1). [PMID: 32003321]
- Desai SK, and Kenney LJ. Switching Lifestyles Is an in vivo Adaptive Strategy of Bacterial Pathogens. Front Cell Infect Microbiol 2019; 9:421. [PMID: 31921700]
- Desai SK, Padmanabhan A, Harshe S, Zaidel-Bar R, and Kenney LJ. Salmonella biofilms program innate immunity for persistence in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 2019;. [PMID: 31160462]
- Ottemann KM, and Kenney LJ. Editorial overview: Host-pathogen interactions: bacteria. Curr. Opin. Microbiol. 2019; 47:iii-v. [PMID: 31138403]