What is Serum Response Factor?
Serum response factor (SRF) is a transcription factor that plays a key role in the transduction of mechanical signals from cytoplasmic actin and the extracellular environment, to the cell nucleus. It is highly conserved from yeast to humans and plays a role in a large number of mechanotransduction pathways. It has been estimated that the transcription of as many as 300 genes is under the control of SRF signalling [1], and of these, more than 200 are directly targeted by the protein [2].
With multiple pathways interacting or converging on SRF, it is considered central to mechanotransduction. NF-kB, zyxin/paxillin, integrin, E-cadherin, Wnt, TGFβ signaling pathways, all represent upstream components of SRF signaling, and as a result, SRF regulates actin-related genes, muscle type-specific genes (i.e., cardiac, smooth, skeletal), as well as genes related to cell survival and apoptosis.
SRF is therefore integral to a vast number of cellular processes, from cell-proliferation, to muscle differentiation and development. It is also important in processes directly related to cytoskeleton dynamics, including cell adhesion and cell migration (reviewed in [3][1]).
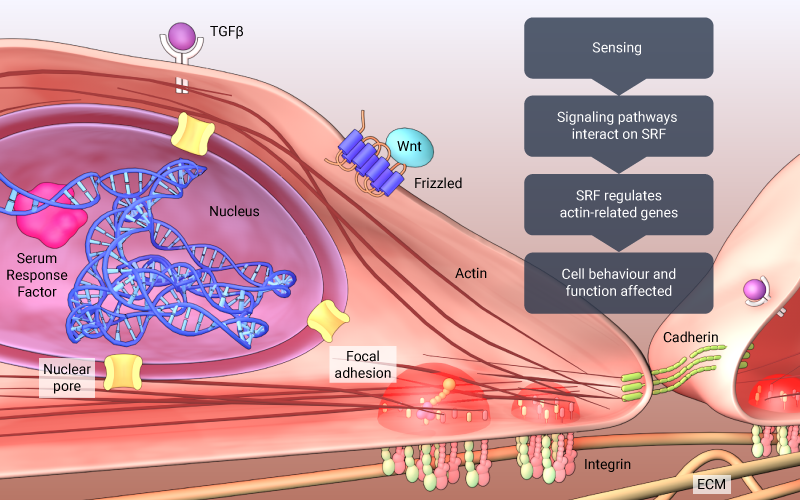
Multiple pathways interact or converge on SRF, which regulates actin-related genes, muscle type-specific genes, and other genes related to cell survival and apoptosis.
A central role in many pathways
SRF activation is directly correlated to cytoskeleton dynamics, and in particular, the levels of globular- and filamentous-actin (G- & F-actin) in the cytoplasm, which are determined by the level of actin filament assembly and disassembly, or actin treadmilling. Studies that over expressed actin mutants, suggest that it is the concentration of free G-actin, rather than the G:F-actin ratio, that directly affects SRF activation [4].
Importantly, SRF does not directly interact with actin, and is instead activated by one of two classes of cofactors. Specifically, myocardin-related transcription factors (MRTFs), including myocardin itself [5][6], and ternary complex factors (TCFs) [7][8] . The former is dependent on signaling from the Rho-actin pathway [5], whereas TCF requires phosphorylation by the MAP kinase pathway. Importantly, these two cofactors compete with each other for mutually exclusive interaction with SRF [8]. SRF localizes to the nucleus in homodimeric form, and once activated by either an MRTF or TCF, will bind with high affinity to the palindromic DNA sequence CC(A/T)2A(A/T)3GG, which is present in the promoter region of each target gene. This sequence is also known as a CArG-box, a C A-rich G –box, or an SRE serum response element [9][10].
Despite the large number of SRF target genes, they can generally be grouped into a small number of groups based on their function. Those genes that encode proteins that are involved in the G0-G1 transition of the cell cycle are mostly regulated by the Ras pathway, and in these cases SRF will be activated by a TCF. [11][12]. However, genes which encode muscle specific proteins involved in contractile functions, cell motility and actin dynamics seem to be regulated by the Rho-MRTF mediated activation of SRF[1]. Actin-related SRF-regulated gene products can be further grouped into structural proteins (e.g., actin, dystrophin, myosin, vinculin), effectors of actin turnover (e.g., cofilin, gelsolin) and regulators of actin dynamics (e.g., talin, filamin) [1].
A role in mechanobiology
With the CArG-box present in the promoters of many muscle development and growth related genes [13][14][15] and with a fundamental role in the regulation of genes related to the actin cytoskeleton [16][17][18], SRF is integral to the cells ability to sense and respond to mechanical cues from its environment.
SRF-signaling has been implicated as a major pathway in the direct regulation of genes responsible for stem cell fate regulation [19][20][21], rigidity sensing [21], inflammatory migration of immune cells [22] and morphogenesis [23].
Conneli et al. [24] have shown that keratinocytes become spread when grown on stiffer surfaces, and exhibit an increase in the formation of mature focal adhesions with a rapid reorganization of actin filaments. When grown on softer surfaces however, the cells round up, and dense cortical actin network is formed. This ultimately depletes the pool of free cellular G-actin releasing MRTFs, and reduces SRF signaling. The end result is terminal differentiation of the cell[19][20]
A review by Taylor and Halene suggest a de novo synthesis of actin and other cytoskeletal proteins is mediated by SRF signaling during the trans-epithelial migration of immune cells [22].
Abnormalities observed in the cytoskeleton of Srf-null embryonic stem cells [25] suggest that SRF is an important regulator of cytoskeletal dynamics. Park et al. have shown that smooth muscle cells (SMC) specific Srf knock-out is embryonic lethal in mice due to severe defects in gastrointestinal and cardiac developments[23].
References
- Olson EN, and Nordheim A. Linking actin dynamics and gene transcription to drive cellular motile functions. Nat. Rev. Mol. Cell Biol. 2010; 11(5):353-65. [PMID: 20414257]
- Cooper SJ, Trinklein ND, Nguyen L, and Myers RM. Serum response factor binding sites differ in three human cell types. Genome Res. 2007; 17(2):136-44. [PMID: 17200232]
- Posern G, and Treisman R. Actin’ together: serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol. 2006; 16(11):588-96. [PMID: 17035020]
- Posern G, Sotiropoulos A, and Treisman R. Mutant actins demonstrate a role for unpolymerized actin in control of transcription by serum response factor. Mol. Biol. Cell 2002; 13(12):4167-78. [PMID: 12475943]
- Miralles F, Posern G, Zaromytidou A, and Treisman R. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell 2003; 113(3):329-42. [PMID: 12732141]
- Wang Z, Wang D, Pipes GCT, and Olson EN. Myocardin is a master regulator of smooth muscle gene expression. Proc. Natl. Acad. Sci. U.S.A. 2003; 100(12):7129-34. [PMID: 12756293]
- Vickers ER, Kasza A, Kurnaz IA, Seifert A, Zeef LAH, O’donnell A, Hayes A, and Sharrocks AD. Ternary complex factor-serum response factor complex-regulated gene activity is required for cellular proliferation and inhibition of apoptotic cell death. Mol. Cell. Biol. 2004; 24(23):10340-51. [PMID: 15542842]
- Wang Z, Wang D, Hockemeyer D, McAnally J, Nordheim A, and Olson EN. Myocardin and ternary complex factors compete for SRF to control smooth muscle gene expression. Nature 2004; 428(6979):185-9. [PMID: 15014501]
- Pellegrini L, Tan S, and Richmond TJ. Structure of serum response factor core bound to DNA. Nature 1995; 376(6540):490-8. [PMID: 7637780]
- Treisman R. Identification of a protein-binding site that mediates transcriptional response of the c-fos gene to serum factors. Cell 1986; 46(4):567-74. [PMID: 3524858]
- Shaw PE, Schröter H, and Nordheim A. The ability of a ternary complex to form over the serum response element correlates with serum inducibility of the human c-fos promoter. Cell 1989; 56(4):563-72. [PMID: 2492906]
- Buchwalter G, Gross C, and Wasylyk B. Ets ternary complex transcription factors. Gene 2004; 324:1-14. [PMID: 14693367]
- Miano JM. Serum response factor: toggling between disparate programs of gene expression. J. Mol. Cell. Cardiol. 2003; 35(6):577-93. [PMID: 12788374]
- Treisman R. Journey to the surface of the cell: Fos regulation and the SRE. EMBO J. 1995; 14(20):4905-13. [PMID: 7588619]
- Spencer JA, and Misra RP. Expression of the serum response factor gene is regulated by serum response factor binding sites. J. Biol. Chem. 1996; 271(28):16535-43. [PMID: 8663310]
- Philippar U, Schratt G, Dieterich C, Müller JM, Galgóczy P, Engel FB, Keating MT, Gertler F, Schüle R, Vingron M, and Nordheim A. The SRF target gene Fhl2 antagonizes RhoA/MAL-dependent activation of SRF. Mol. Cell 2004; 16(6):867-80. [PMID: 15610731]
- Selvaraj A, and Prywes R. Expression profiling of serum inducible genes identifies a subset of SRF target genes that are MKL dependent. BMC Mol. Biol. 2004; 5:13. [PMID: 15329155]
- Sun Q, Chen G, Streb JW, Long X, Yang Y, Stoeckert CJ, and Miano JM. Defining the mammalian CArGome. Genome Res. 2005; 16(2):197-207. [PMID: 16365378]
- Costa P, Almeida FVM, and Connelly JT. Biophysical signals controlling cell fate decisions: how do stem cells really feel? Int. J. Biochem. Cell Biol. 2012; 44(12):2233-7. [PMID: 22982240]
- Lv H, Li L, Sun M, Zhang Y, Chen L, Rong Y, and Li Y. Mechanism of regulation of stem cell differentiation by matrix stiffness. Stem Cell Res Ther 2015; 6:103. [PMID: 26012510]
- O’Connor JW, Riley PN, Nalluri SM, Ashar PK, and Gomez EW. Matrix Rigidity Mediates TGFβ1-Induced Epithelial-Myofibroblast Transition by Controlling Cytoskeletal Organization and MRTF-A Localization. J. Cell. Physiol. 2015; 230(8):1829-39. [PMID: 25522130]
- Taylor A, and Halene S. The regulatory role of serum response factor pathway in neutrophil inflammatory response. Curr. Opin. Hematol. 2015; 22(1):67-73. [PMID: 25402621]
- Park C, Lee MY, Park PJ, Ha SE, Berent RM, Fuchs R, Miano JM, Becker LS, Sanders KM, and Ro S. Serum Response Factor Is Essential for Prenatal Gastrointestinal Smooth Muscle Development and Maintenance of Differentiated Phenotype. J Neurogastroenterol Motil 2015; 21(4):589-602. [PMID: 26424044]
- Connelly JT, Gautrot JE, Trappmann B, Tan DW, Donati G, Huck WTS, and Watt FM. Actin and serum response factor transduce physical cues from the microenvironment to regulate epidermal stem cell fate decisions. Nat. Cell Biol. 2010; 12(7):711-8. [PMID: 20581838]
- Schratt G, Philippar U, Berger J, Schwarz H, Heidenreich O, and Nordheim A. Serum response factor is crucial for actin cytoskeletal organization and focal adhesion assembly in embryonic stem cells. J. Cell Biol. 2002; 156(4):737-50. [PMID: 11839767]


