Alpha-actinin
α-actinin is an actin-binding protein [1] and component of the actin crosslinking functional modules; it lacks G-actin binding activity and lacks actin initiation/nucleation activity [2]. α-actinin is an important organizer of the cytoskeleton that belongs to the spectrin superfamily (which includes spectrin, dystrophin, and related homologues). α-actinin is present in a number of diverse organisms including protists, invertebrates, and birds; mammals have at least four α-actinin genes that together account for 6 different α-actinin proteins whose expression profile is tissue specific (reviewed in [3]).
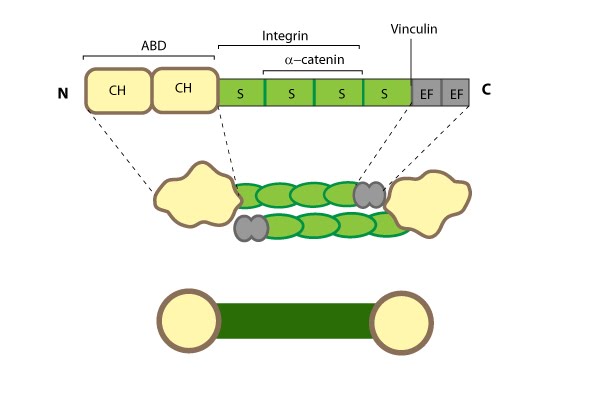
This schematic diagram illustrates the molecular organization of α-actinin (reviewed in [PMID: 18488141]) and provides examples for how α-actinin is represented in figures throughout this resource. Relevant domains/regions that are believed to be important for actin binding and protein-protein interactions are highlighted (actin binding domain (ABD) [PMID:3733725], β-integrin [PMID:2116421, 15721583], α-catenin [PMID:9152027] and vinculin [PMID:15988023, 8037676]).
The basic organization of the ABDs is quite similar in other members of the α-actinin superfamily such as filamin, fimbrin, and parvin [8]. α-actinin binds F-actin [9] and other molecules such as phophatidylinositol-bisphosphate (PIP2) [10], cell adhesion proteins (e.g. *integrins [11][12]) and signaling enzymes (e.g. PI3K [13]). It also interacts with *vinculin through the 4th S repeat [14] and this interaction acts as a regulatory switch in adherens junctions [15]. Intramolecular contacts that sterically prevent α-actinin from interacting with actin filaments and integrins are relieved by PIP2 binding to the ABD [16] and this regulates α-actinin dynamics [17].
Dimerization of α-actinin via the rod domain is also essential for crosslinking actin [18] and for binding to other proteins (e.g. *zyxin) [19], therefore, a dimer has functional domains at both ends [20]; this organization allows them to bind to adjacent actin filaments [21]. The smaller size of the α-actinin dimer combined with flexible hinges at the ABDs, makes α-actinin a versatile actin crosslinker capable of forming variable orientations and angles between actin filaments, as well as forming tighter bridges between filaments (such as those found in actin bundles) (reviewed in [3]).
α-actinin localization and function
α-actinin primarily influences the cohesiveness and mechanics of the cytoskeleton by cross-linking actin filaments and other cytoskeleton components to create a scaffold that imparts stability and forms a bridge between the cytoskeleton and signaling pathways. α-actinin interacts with numerous (~30) components in the cell (reviewed in [22]) and certain α-actinin isoforms (and related proteins) appear to be active in the nucleus (reviewed in [23]). α-actinin is mainly found at the leading edge of migrating cells and it is an important component of adhesion modules [24]. Dendritic spines are also rich in α-actinin and it appears to play a role in neuritic outgrowth [25]. Lastly, α-actinin is believed to be the primary crosslinking protein in stress fibers [26] and it plays a major role in the maturation of focal adhesions [27]. Localization of α-actinin to the plasma membrane is controlled by a number of interactions with membrane lipids and transmembrane receptors (reviewed in [2]). For example, binding of PIP2 at the plasma membrane causes a conformational change in the CaM-like domain that subsequently increases α-actinin’s affinity for actin and its ability to interact with other cytoskeletal components (e.g. titin [28]) (reviewed in [3]).
References
- Maruyama K, and Ebashi S. Alpha-actinin, a new structural protein from striated muscle. II. Action on actin. J. Biochem. 1965; 58(1):13-9. [PMID: 5857097]
- Ohtaki T, Tsukita S, Mimura N, Tsukita S, and Asano A. Interaction of actinogelin with actin. No nucleation but high gelation activity. Eur. J. Biochem. 1985; 153(3):609-20. [PMID: 4076192]
- Sjöblom B, Salmazo A, and Djinović-Carugo K. Alpha-actinin structure and regulation. Cell. Mol. Life Sci. 2008; 65(17):2688-701. [PMID: 18488141]
- Burridge K, and Feramisco JR. Non-muscle alpha actinins are calcium-sensitive actin-binding proteins. Nature 1981; 294(5841):565-7. [PMID: 7312045]
- Bennett JP, Zaner KS, and Stossel TP. Isolation and some properties of macrophage alpha-actinin: evidence that it is not an actin gelling protein. Biochemistry 1984; 23(21):5081-6. [PMID: 6498177]
- Duhaiman AS, and Bamburg JR. Isolation of brain alpha-actinin. Its characterization and a comparison of its properties with those of muscle alpha-actinins. Biochemistry 1984; 23(8):1600-8. [PMID: 6722113]
- Landon F, Gache Y, Touitou H, and Olomucki A. Properties of two isoforms of human blood platelet alpha-actinin. Eur. J. Biochem. 1985; 153(2):231-7. [PMID: 2934249]
- Olski TM, Noegel AA, and Korenbaum E. Parvin, a 42 kDa focal adhesion protein, related to the alpha-actinin superfamily. J. Cell. Sci. 2001; 114(Pt 3):525-38. [PMID: 11171322]
- Mimura N, and Asano A. Isolation and characterization of a conserved actin-binding domain from rat hepatic actinogelin, rat skeletal muscle, and chicken gizzard alpha-actinins. J. Biol. Chem. 1986; 261(23):10680-7. [PMID: 3733725]
- Fukami K, Sawada N, Endo T, and Takenawa T. Identification of a phosphatidylinositol 4,5-bisphosphate-binding site in chicken skeletal muscle alpha-actinin. J. Biol. Chem. 1996; 271(5):2646-50. [PMID: 8576235]
- Otey CA, Pavalko FM, and Burridge K. An interaction between alpha-actinin and the beta 1 integrin subunit in vitro. J. Cell Biol. 1990; 111(2):721-9. [PMID: 2116421]
- Kelly DF, and Taylor KA. Identification of the beta1-integrin binding site on alpha-actinin by cryoelectron microscopy. J. Struct. Biol. 2005; 149(3):290-302. [PMID: 15721583]
- Shibasaki F, Fukami K, Fukui Y, and Takenawa T. Phosphatidylinositol 3-kinase binds to alpha-actinin through the p85 subunit. Biochem. J. 1994; 302 ( Pt 2):551-7. [PMID: 8093010]
- McGregor A, Blanchard AD, Rowe AJ, and Critchley DR. Identification of the vinculin-binding site in the cytoskeletal protein alpha-actinin. Biochem. J. 1994; 301 ( Pt 1):225-33. [PMID: 8037676]
- Bois PRJ, Borgon RA, Vonrhein C, and Izard T. Structural dynamics of alpha-actinin-vinculin interactions. Mol. Cell. Biol. 2005; 25(14):6112-22. [PMID: 15988023]
- Young P, and Gautel M. The interaction of titin and alpha-actinin is controlled by a phospholipid-regulated intramolecular pseudoligand mechanism. EMBO J. 2000; 19(23):6331-40. [PMID: 11101506]
- Fraley TS, Pereira CB, Tran TC, Singleton C, and Greenwood JA. Phosphoinositide binding regulates alpha-actinin dynamics: mechanism for modulating cytoskeletal remodeling. J. Biol. Chem. 2005; 280(15):15479-82. [PMID: 15710624]
- Djinović-Carugo K, Young P, Gautel M, and Saraste M. Structure of the alpha-actinin rod: molecular basis for cross-linking of actin filaments. Cell 1999; 98(4):537-46. [PMID: 10481917]
- Li B, and Trueb B. Analysis of the alpha-actinin/zyxin interaction. J. Biol. Chem. 2001; 276(36):33328-35. [PMID: 11423549]
- Ylänne J, Scheffzek K, Young P, and Saraste M. Crystal structure of the alpha-actinin rod reveals an extensive torsional twist. Structure 2001; 9(7):597-604. [PMID: 11470434]
- Goli DE, Suzuki A, Temple J, and Holmes GR. Studies on purified -actinin. I. Effect of temperature and tropomyosin on the -actinin-F-actin interaction. J. Mol. Biol. 1972; 67(3):469-88. [PMID: 5045308]
- Otey CA, and Carpen O. Alpha-actinin revisited: a fresh look at an old player. Cell Motil. Cytoskeleton 2004; 58(2):104-11. [PMID: 15083532]
- Uribe R, and Jay D. A review of actin binding proteins: new perspectives. Mol. Biol. Rep. 2007; 36(1):121-5. [PMID: 17939058]
- Knight B, Laukaitis C, Akhtar N, Hotchin NA, Edlund M, and Horwitz AR. Visualizing muscle cell migration in situ. Curr. Biol. 2000; 10(10):576-85. [PMID: 10837222]
- Nyman-Huttunen H, Tian L, Ning L, and Gahmberg CG. alpha-Actinin-dependent cytoskeletal anchorage is important for ICAM-5-mediated neuritic outgrowth. J. Cell. Sci. 2006; 119(Pt 15):3057-66. [PMID: 16820411]
- Zaidel-Bar R, Itzkovitz S, Ma’ayan A, Iyengar R, and Geiger B. Functional atlas of the integrin adhesome. Nat. Cell Biol. 2007; 9(8):858-67. [PMID: 17671451]
- Choi CK, Vicente-Manzanares M, Zareno J, Whitmore LA, Mogilner A, and Horwitz AR. Actin and alpha-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner. Nat. Cell Biol. 2008; 10(9):1039-50. [PMID: 19160484]
- Young P, Ferguson C, Bañuelos S, and Gautel M. Molecular structure of the sarcomeric Z-disk: two types of titin interactions lead to an asymmetrical sorting of alpha-actinin. EMBO J. 1998; 17(6):1614-24. [PMID: 9501083]


