Silencing and Anti-Silencing in Salmonella Pathogenesis
Characterizing the binding properties of the H-NS homologue StpA (Lim et al., 2012b) and H-NS mutants that are unable to polymerize
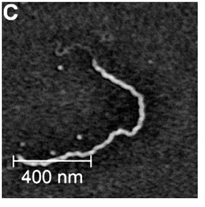
AFM image of H-NS bound to DNA from (Liu et al., 2010). At low Mg2+ concentration, H-NS binds to DNA in a polymerization mode, which leads to stiffening and elongation. This is the form that is de-repressed by SsrB, leading to transcriptional activation (Liu et al., 2010, Walthers et al., 2011).
There are many important questions related to the control of gene expression, since the molecular mechanisms are not well understood. When Salmonella resides in a macrophage vacuole, the EnvZ/OmpR two-component system activates transcription of the SsrA/B system located on pathogenicity island 2 (SPI-2), in response to acid stress (Fig. 4). The response regulator SsrB activates expression of genes encoded within and outside of SPI-2, which are required for systemic infection. SsrB binds upstream of the sifA, sifB, and sseJ effector genes and directly regulates transcription (Walthers et al., 2011).
SsrB also relieves gene silencing by the nucleoid protein H-NS. Single molecule experiments with magnetic tweezers demonstrated that H-NS has two binding modes that are influenced by Mg2+ or other divalent cations (Liu et al., 2010). SsrB displaces H-NS from DNA only when it is bound in a polymerization (stiffening) mode and not when H-NS is bound to DNA in the bridging mode (Walthers et al., 2011). Thus, in contrast to previous views, the polymerization-binding mode of H-NS is the relevant form for counter-silencing by SsrB (Fig. 5). Our results reveal that response regulators can directly activate transcription and also relieve H-NS silencing. These experiments are part of an ongoing collaboration with Dr. Jie Yan in the Department of Physics at NUS. We have recently characterized the binding properties of the H-NS homologue StpA (Lim et al., 2012b) as well as H-NS mutants that are unable to polymerize (Lim et al., 2012a).
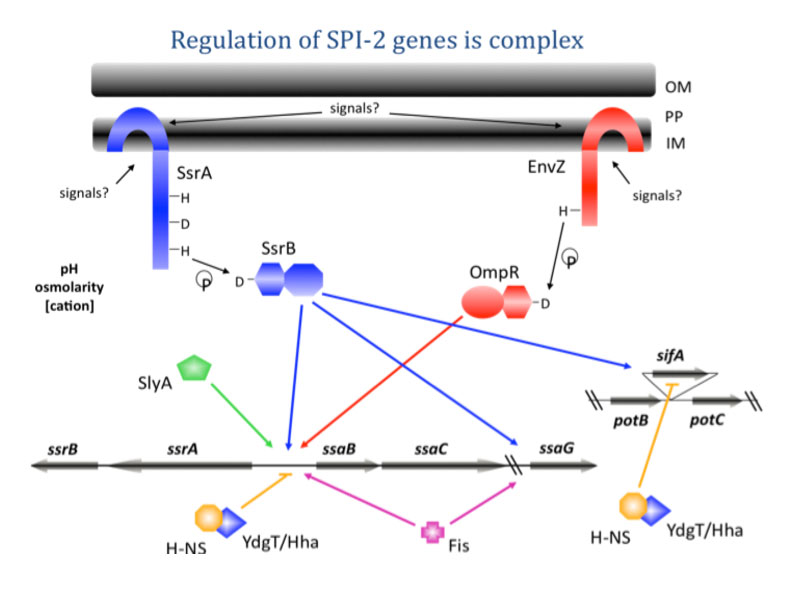
A summary of our current view of OmpR/EnvZ regulation of ssrA/B. EnvZ phosphorylates OmpR (step 1) in response to an unknown stress (acid pH?). OmpR~P binds with high affinity upstream of ssrA and activates transcription (step 2). SsrA phosphorylates SsrB (step 3) and SsrB~P activates transcription of genes both within and outside of SPI-2 (step 4). OmpR also regulates additional genes such as sseI (Feng et al., 2004) and sifA (Pickard et al., 1994).
Researchers: Priyanka Das, Stuti Desai
Lab: Linda J Kenney Lab


