What is Myosin?
An Introduction to the Myosin Superfamily of Proteins
Myosin I has unique tail domain(s) relative to other myosin members which allows myosin I to bind to membrane lipids or to more than one actin filament at a time (see panel ‘A’ in Figure below). Myosin I is primarily involved in intracellular organization, but it also forms a critical component of small cell surface projections in intestinal cells.
Myosin II can form higher order assemblies via the extended coiled-coil domains in the heavy chains. For example, the long coiled-coil domains of myosin II interact with the coiled-coiled domains of adjacent myosin II molecules, followed by additional tail-tail interactions with other myosin II assemblies. The resulting myosin II bundle (aka ‘”thick filament”) has several hundred myosin heads oriented in opposite directions at the two ends of the filament. Concerted ATP hydrolysis and movement of the myosin heads along adjacent actin filaments generates a sliding motion that results in shortening or contraction of the interlinked actin filaments (see arrows in panel ‘B’ in Figure below). The action of the actin-myosin system generates forces against the interlinked cytoskeleton network to influence processes such as cell signaling, adhesion, movement, polarity, and cell fate (see “contractile bundle” in the main glossary) [1][2][3] (reviewed in [4]). Myosin II is also a critical component of stress fibers and the contractile ring that separates two cells during cell division. For studies which investigate cell contraction and motility, contractile force generated by myosin II can be inhibited using small molecules such as blebbistatin [5] and 2,3-butanedione monoxime (BDM) [6].
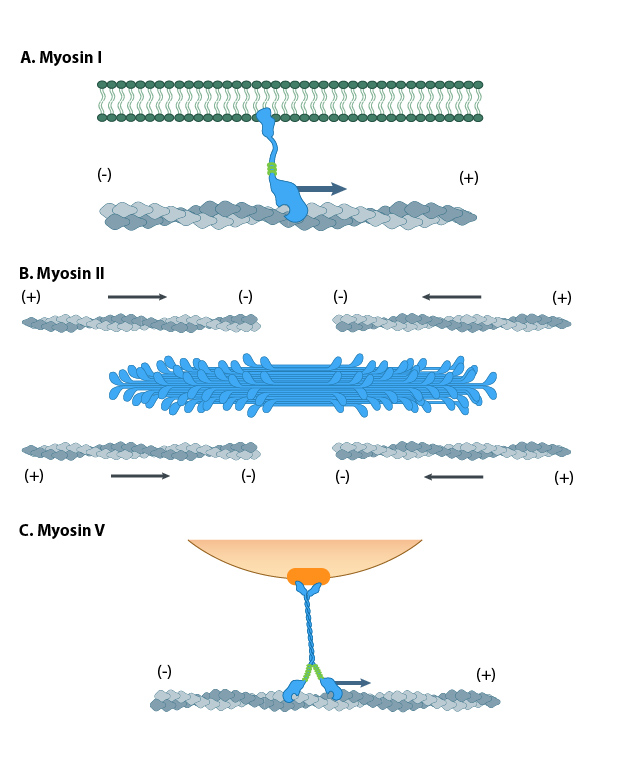
Different motor protein functions of the myosin family. A. Myosin I can bind to membrane lipids. B. Bundles of Myosin II slide along the actin cytoskeleton network to drive actomyosin contractility. C. Myosin V transports cargo by ‘walking’ l along actin filaments.
Myosin V and Myosin VI In non-muscle cells, actin filaments form an internal track system for cargo transport that is powered by motor proteins such as myosin V and myosin VI ( see panel ‘C’ in Figure below); these myosins use the energy from ATP hydrolysis to transport cargo (such as attached vesicles and organelles) at rates much faster than diffusion. Myosin V contains more light chains and a longer ‘lever arm’ relative to myosin II, which allows myosin V to move in larger steps along the actin filaments (reviewed in [7]).
Myosin V may also colocalize with F-actin bundles. The distribution of myosin V in growth cones is consistent with the role for this myosin in tension production by growth cones. Myosin V may influence the filopodial extension rate by pushing the plasma membrane and creating space for G-actin subunit assembly onto the barbed ends of actin filaments [8][9].
Myosin VII and Myosin X are important for filopodial assembly and dynamics [10][11]. Myosin VII is believed to influence the assembly/disassembly of adhesion proteins at the filopodial tip [10] as well as play a role in filopodial extension events. Myosin X activity also influences the filopodia number and overall length with calmodulin-like protein (CLP) modulating this activity by stabilizing myosin X [12]. Myosin X influences transport of materials along filopodial shafts using an ATP-dependent ‘walking’ mechanism. Myosin X binds cell surface receptors, cytoskeleton, Ena/VASP proteins, and membrane phospholipids [10][13][14][15]. Myosin X also has a striking distribution at the tips of filopodia and disrupting its function disrupts filopodium formation [16][17].
Myosin motor protein structure
Several myosin isoforms have been found in eukaryotes, each differing in the type of heavy and light chains they are composed of. All myosins are composed of a diverse ‘tail’ domain at their carboxy terminus and an evolutionarily conserved globular ‘head’ domain at their amino terminus. The diverse ‘tails’ of different myosin isoforms bind specific substrates or cargo, whilst their conserved ‘heads’ contain sites for ATP binding [18], F-actin binding and force generation (i.e. motor domains) (reviewed in [19][20]).
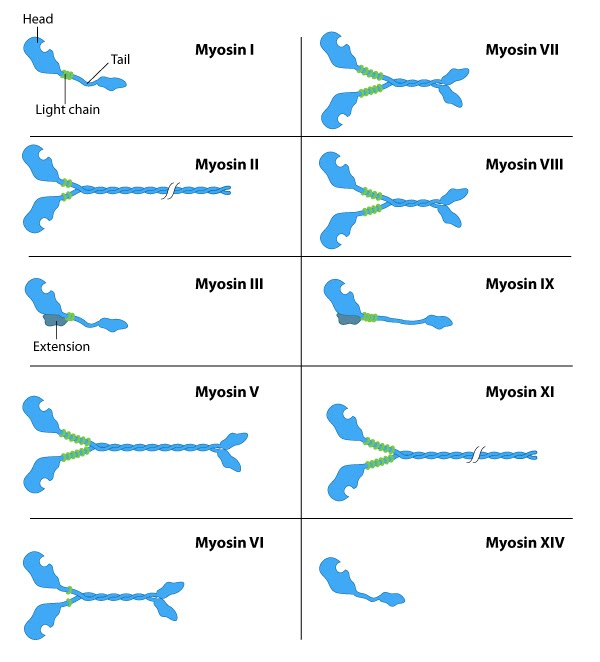
All myosins share a motor domain on their heavy chains at the amino-terminus (the ‘head’ domain), but they differ considerably at their carboxy-terminus (the ‘tail’ domain). A few myosin types also have an amino-terminal extension. The number of light chains varies considerably between myosin types and certain myosins exist as dimers. Myosins that form dimers have two motor domains, and the number of light chains can influence the “lever arm” length between the myosin heads – this regulates the length of the myosin ‘powerstroke’ and the distance the myosin can travel along the actin filament in a single round of ATP hydrolysis (see also ‘myosin powerstroke’).
All myosins bind to actin filaments via a globular ‘head’ domain located at the end of the heavy chains. Actin binding to this region increases the ATPase activity of myosins (reviewed in [2][4]. Some myosins have a single heavy chain and contact actin filaments at only one site, while other myosin isoforms have two heavy chains and contact actin filaments at two sites. Myosin II is the only family member that can form polymeric assemblies [20]).
The number of light chains influences the length of the “lever arm” or “neck region” and therefore the “step size” of different myosin types [21]. Myosin V contains more light chains relative to myosin II and so myosin V moves in larger steps along actin filaments after an equivalent round of ATP hydrolysis (reviewed in [22]).
Myosin motors move along actin filaments in defined directions. With the exception of myosin VI, which moves towards the pointed end, all myosins move towards the barbed end. Most actin filaments have the barbed end directed towards the plasma membrane and the pointed end towards the interior. This arrangement allows certain myosins (e.g. myosin V) to function primarily for cargo export, while myosin VI acts as the major motor protein for import. Myosin II is commonly associated with retraction fibers and retrograde actin flow at the pointed end of actin filaments. All non-muscle cells use contractile bundles containing myosin II to generate forces that promote the assembly of actin filaments.
Although most myosins function as motor proteins in the cytoplasm, some species of myosin are localized to, and function in, the nucleus. Nuclear Myosin I (NMI), myosin II, myosin V, myosin VI, myosin XVIB and myosin XVIIIB have all been found in the nucleus [23][24][25], with NMI being the most extensively studied.
References
- Shelden E, and Knecht DA. Mutants lacking myosin II cannot resist forces generated during multicellular morphogenesis. J. Cell. Sci. 1995; 108 ( Pt 3):1105-15. [PMID: 7622597]
- Vicente-Manzanares M, Zareno J, Whitmore L, Choi CK, and Horwitz AF. Regulation of protrusion, adhesion dynamics, and polarity by myosins IIA and IIB in migrating cells. J. Cell Biol. 2007; 176(5):573-80. [PMID: 17312025]
- Even-Ram S, Doyle AD, Conti MA, Matsumoto K, Adelstein RS, and Yamada KM. Myosin IIA regulates cell motility and actomyosin-microtubule crosstalk. Nat. Cell Biol. 2007; 9(3):299-309. [PMID: 17310241]
- Cai Y, and Sheetz MP. Force propagation across cells: mechanical coherence of dynamic cytoskeletons. Curr. Opin. Cell Biol. 2009; 21(1):47-50. [PMID: 19208463]
- Straight AF, Cheung A, Limouze J, Chen I, Westwood NJ, Sellers JR, and Mitchison TJ. Dissecting temporal and spatial control of cytokinesis with a myosin II Inhibitor. Science 2003; 299(5613):1743-7. [PMID: 12637748]
- Zhao L, Naber N, and Cooke R. Muscle cross-bridges bound to actin are disordered in the presence of 2,3-butanedione monoxime. Biophys. J. 1995; 68(5):1980-90. [PMID: 7612840]
- Trybus KM. Myosin V from head to tail. Cell. Mol. Life Sci. 2008; 65(9):1378-89. [PMID: 18239852]
- Sheetz MP, Wayne DB, and Pearlman AL. Extension of filopodia by motor-dependent actin assembly. Cell Motil. Cytoskeleton 1992; 22(3):160-9. [PMID: 1423662]
- Wang FS, Wolenski JS, Cheney RE, Mooseker MS, and Jay DG. Function of myosin-V in filopodial extension of neuronal growth cones. Science 1996; 273(5275):660-3. [PMID: 8662560]
- Berg JS, and Cheney RE. Myosin-X is an unconventional myosin that undergoes intrafilopodial motility. Nat. Cell Biol. 2002; 4(3):246-50. [PMID: 11854753]
- Tuxworth RI, Weber I, Wessels D, Addicks GC, Soll DR, Gerisch G, and Titus MA. A role for myosin VII in dynamic cell adhesion. Curr. Biol. 2001; 11(5):318-29. [PMID: 11267868]
- Bennett RD, Mauer AS, and Strehler EE. Calmodulin-like protein increases filopodia-dependent cell motility via up-regulation of myosin-10. J. Biol. Chem. 2006; 282(5):3205-12. [PMID: 17130134]
- Tokuo H, and Ikebe M. Myosin X transports Mena/VASP to the tip of filopodia. Biochem. Biophys. Res. Commun. 2004; 319(1):214-20. [PMID: 15158464]
- Zhang H, Berg JS, Li Z, Wang Y, Lång P, Sousa AD, Bhaskar A, Cheney RE, and Strömblad S. Myosin-X provides a motor-based link between integrins and the cytoskeleton. Nat. Cell Biol. 2004; 6(6):523-31. [PMID: 15156152]
- Zhu X, Wang C, Dai P, Xie Y, Song N, Liu Y, Du Q, Mei L, Ding Y, and Xiong W. Myosin X regulates netrin receptors and functions in axonal path-finding. Nat. Cell Biol. 2007; 9(2):184-92. [PMID: 17237772]
- Bohil AB, Robertson BW, and Cheney RE. Myosin-X is a molecular motor that functions in filopodia formation. Proc. Natl. Acad. Sci. U.S.A. 2006; 103(33):12411-6. [PMID: 16894163]
- De Joussineau C, Soulé J, Martin M, Anguille C, Montcourrier P, and Alexandre D. Delta-promoted filopodia mediate long-range lateral inhibition in Drosophila. Nature 2003; 426(6966):555-9. [PMID: 14654840]
- Walker JE, Saraste M, Runswick MJ, and Gay NJ. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982; 1(8):945-51. [PMID: 6329717]
- Hwang W, and Lang MJ. Mechanical design of translocating motor proteins. Cell Biochem. Biophys. 2009; 54(1-3):11-22. [PMID: 19452133]
- Cheney RE, Riley MA, and Mooseker MS. Phylogenetic analysis of the myosin superfamily. Cell Motil. Cytoskeleton 1993; 24(4):215-23. [PMID: 8477454]
- Uyeda TQ, Abramson PD, and Spudich JA. The neck region of the myosin motor domain acts as a lever arm to generate movement. Proc. Natl. Acad. Sci. U.S.A. 1996; 93(9):4459-64. [PMID: 8633089]
- Ruppel KM, and Spudich JA. Structure-function analysis of the motor domain of myosin. Annu. Rev. Cell Dev. Biol. 1996; 12:543-73. [PMID: 8970737]
- Sobczak M, Majewski �, and Redowicz MJ. [Myosins in nucleus]. Postepy Biochem. 2009; 55(2):239-46. [PMID: 19824481]
- Li Q, and Sarna SK. Nuclear myosin II regulates the assembly of preinitiation complex for ICAM-1 gene transcription. Gastroenterology 2009; 137(3):1051-60, 1060.e1-3. [PMID: 19328794]
- Pranchevicius MCS, Baqui MMA, Ishikawa-Ankerhold HC, Lourenço EV, Leão RM, Banzi SR, dos Santos CT, Roque-Barreira MC, Barreira MCR, Espreafico EM, and Larson RE. Myosin Va phosphorylated on Ser1650 is found in nuclear speckles and redistributes to nucleoli upon inhibition of transcription. Cell Motil. Cytoskeleton 2008; 65(6):441-56. [PMID: 18330901]


