How does chromatin condensation keep the nucleus compact?
Packaging of approximately a meter-long DNA duplex within the elastic nucleus determines its spatial 3D architecture and the genomic accessibility of tissue-specific transcriptional programs. Apart from several biochemical processes, there are various forces at play in the chromatin organization, that in turn affect nuclear functions (reviewed in [1]). DNA micromanipulation experiments reveal that it has a persistent length of ~50nm and hence in an entropic configuration will have a radius of gyration ~300um due to electrostatic repelling as a consequence of its negative charge [2][3]. However, compaction of this flexible polymer inside a much smaller nucleus (~10-50um) is possible only because of the positively charged histones and other non-histone proteins that enable condensation, by stabilizing the electrostatic interactions [4][5].
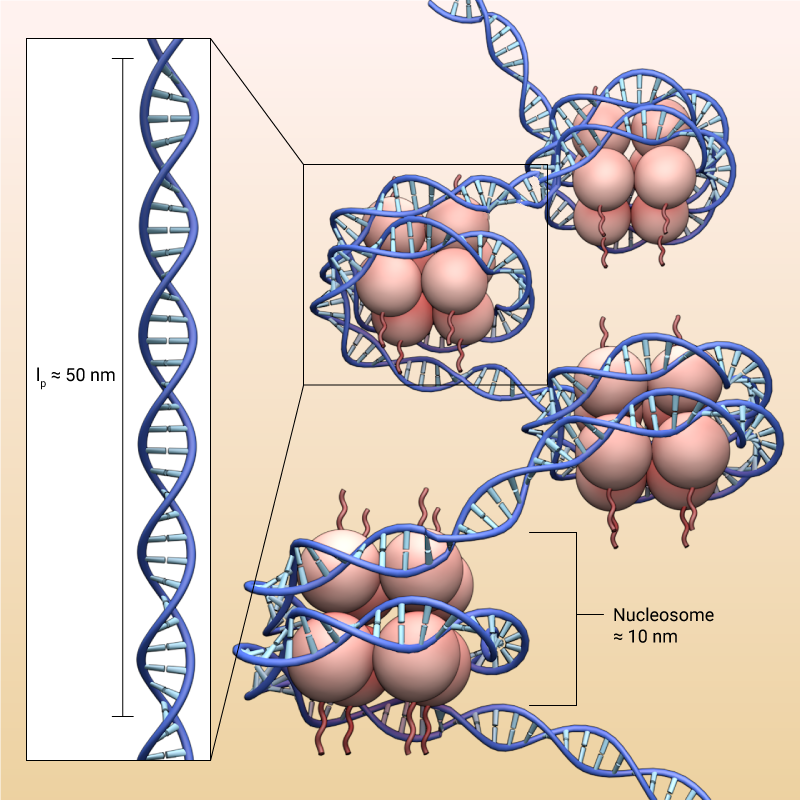
DNA has a persistent length of around 50 nm, but at the same time it wraps tightly around histones forming nucleosomes that are around 10 nm in diameter. This is possible due to the electrostatic attraction between the negatively charged DNA and positively charged histones, and the difference in electrostatic charges between the proteins enable condensation of chromatin.
References
- Shivashankar GV. Mechanosignaling to the cell nucleus and gene regulation. Annu Rev Biophys 2011; 40:361-78. [PMID: 21391812]
- Morton NE. Parameters of the human genome. Proc. Natl. Acad. Sci. U.S.A. 1991; 88(17):7474-6. [PMID: 1881886]
- Bustamante C, Bryant Z, and Smith SB. Ten years of tension: single-molecule DNA mechanics. Nature 2003; 421(6921):423-7. [PMID: 12540915]
- Luger K, and Hansen JC. Nucleosome and chromatin fiber dynamics. Curr. Opin. Struct. Biol. 2005; 15(2):188-96. [PMID: 15837178]
- Marenduzzo D, Micheletti C, and Cook PR. Entropy-driven genome organization. Biophys. J. 2006; 90(10):3712-21. [PMID: 16500976]


