BACTERIAL PATHOGENESIS & SIGNAL TRANSDUCTION LAB
Linda J Kenney’s laboratory employs interdisciplinary approaches to understand environmental signaling in bacteria, host specificity of bacterial pathogens, gene silencing and relief of silencing in Salmonella
Recent Featured Research
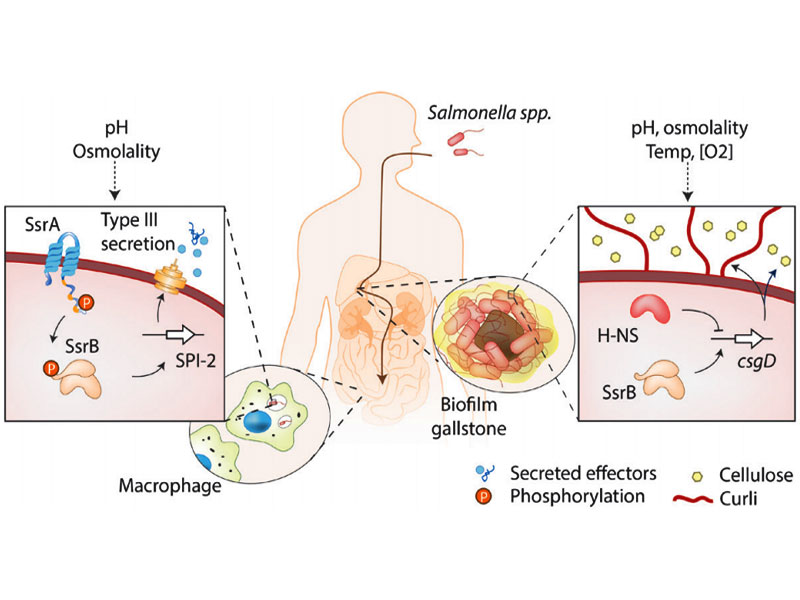
Why Salmonella decide to lay low
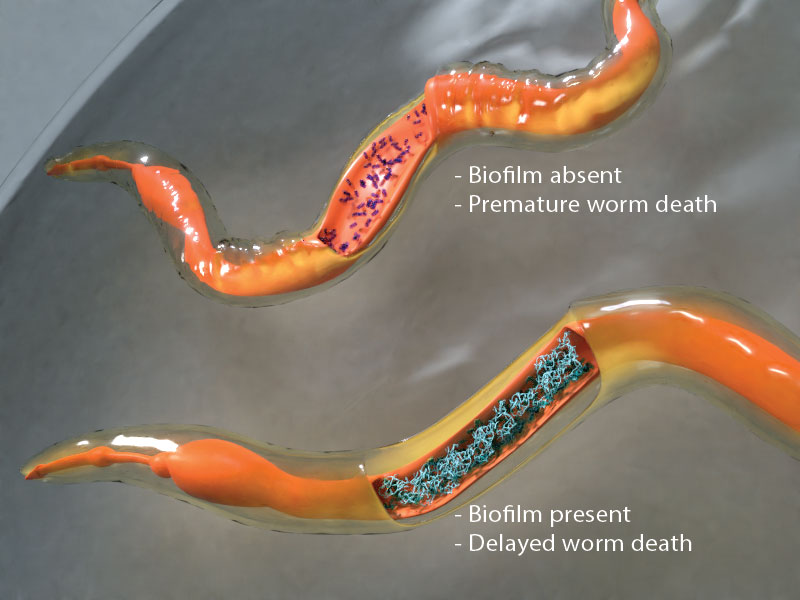
Studying dormant biofilms in live worm hosts
Controlling how bacteria respond to stress
Bacterial self-protection by self-acidification
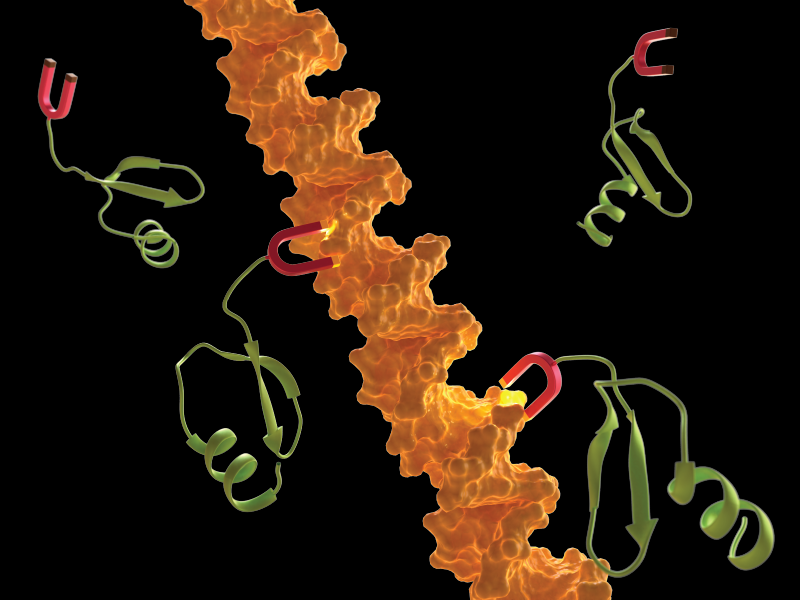
A link to bacterial gene silencing
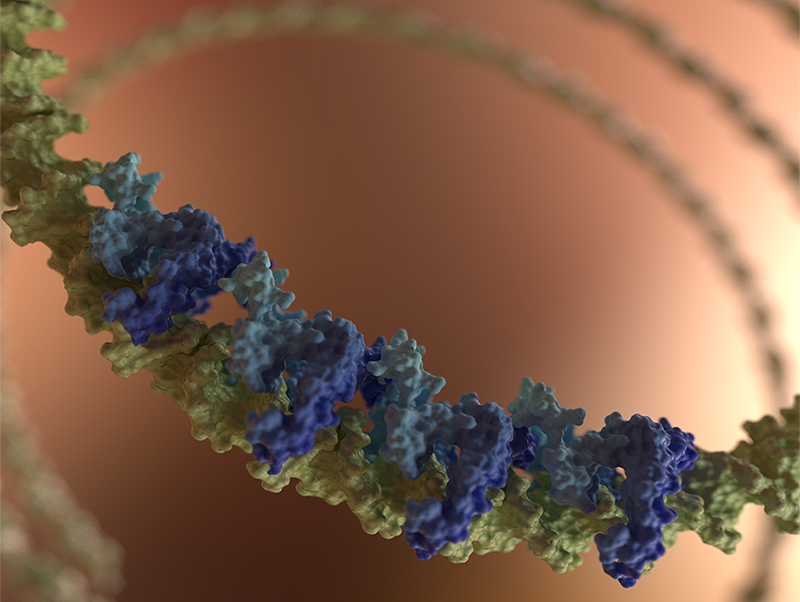
The charged H-NS linker is key to promoting DNA binding
Salmonella Lifestyle Choices
A bacterial molecular switch between virulence or dormancy
H-NS Proteins as Gene Silencers
A gene-silencing nucleoprotein filament formed by H-NS family proteins
Surviving Immune Attack
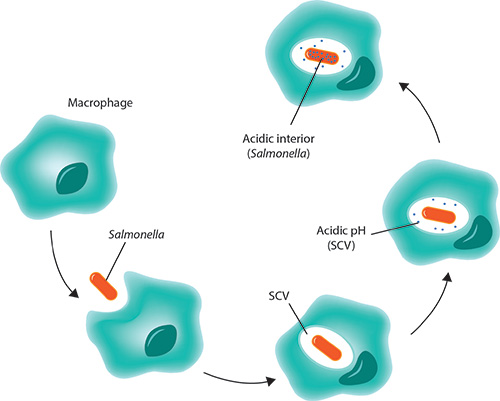
How does Salmonella thrive in the acidic macrophage?
KENNEY LABORATORY RESEARCH
The Kenney laboratory is dedicated to solving important problems in bacterial mechanotransduction related to pathogenesis.
Bacterial pathogenesis involves the programmed changes in gene expression to evade host defense systems through sensing the host environment both physically and chemically.
Research
KENNEY LABORATORY RESEARCH
In bacteria, signal transduction occurs largely via two-component regulatory systems. These systems employ an inner membrane histidine kinase (the sensor) and a cytoplasmic response regulator. Environmental sensing is in most cases directly coupled to regulation of gene expression.
The two components communicate via a series of phosphorylation/ phosphotransfer reactions. The sensor kinase is autophosphorylated by ATP at a conserved histidine residue upon activation by environmental stress. The sensor kinase then transfers the phosphoryl group to a conserved aspartate on the response regulator. Phosphorylation of the response regulator often increases its affinity for DNA leading to transcriptional activation or repression, depending on the mode of action.
KENNEY LABORATORY RESEARCH CONT.
The EnvZ/OmpR system that we study responds to osmotic changes and controls the expression of the outer membrane proteins OmpF and OmpC. OmpR is an important global regulator and plays a major role in regulating virulence genes in many pathogens, including Salmonella, Yersinia, Shigella and E. coli.
A central question in the EnvZ/OmpR two-component system that we study (and an unanswered question in most other sensor kinases as well) is how does a change in the osmolality of the external environment (the signal) result in the differential regulation of outer membrane porin gene expression (the output)?
Current Projects
RECENT WORK
The Kenney laboratory is interested in signal transduction and the regulation of gene expression in prokaryotes. In particular, we are studying the two-component regulatory system EnvZ/OmpR that regulates the expression of outer membrane proteins as well as many other genes.
Our present work focuses on how OmpR activates genes required for systemic infection (located on Salmonella pathogenicity island 2) in Salmonella enterica.
Current Projects

Structural Analysis of SPI-2 needles
Using cryoelectron microscopy to perform structural analysis of the macromolecular complex of the SPI-2 secretory apparatus
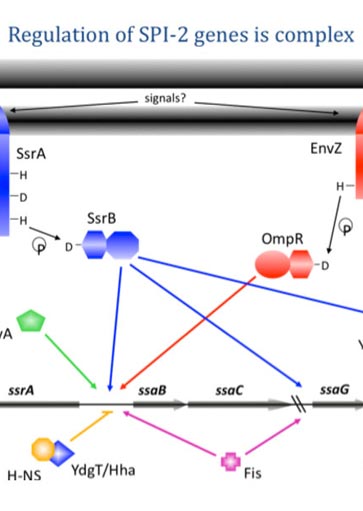
Silencing and Anti-Silencing in Salmonella Pathogenesis
Characterizing the binding properties of the H-NS homologue StpA
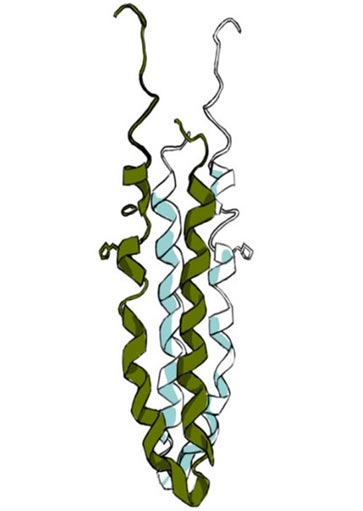
The Mechanism of Osmosensing by EnvZ and other HKs
Mapping conformational changes in EnvZ associated with osmotic stress
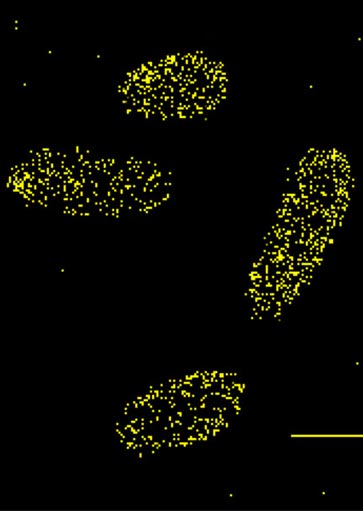
Super-Resolution Microscopy of EnvZ, OmpR, H-NS, and Chromosomal Imaging
Examining OmpR/DNA interactions using PALM
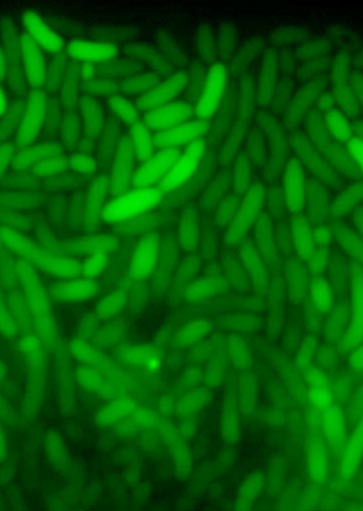
Porin Gene Transcription in Single Cells
Transcription of GFP enables us to see how expression levels change in response to osmolality
Recent Publications
- Fernandez M, Yamanaka Y, Zangoui P, White MA, and Kenney LJ. The sulfur assimilation pathway mitigates redox stress from acidic pH in Salmonella Typhi H58. mBio 2025;:e0046725. [PMID: 40422406]
- Shetty D, and Kenney LJ. A pH-sensitive switch activates virulence in Salmonella. Elife 2023; 12. [PMID: 37706506]
- Oh D, Liu X, Sheetz MP, and Kenney LJ. Small, Dynamic Clusters of Tir-Intimin Seed Actin Polymerization. Small 2023;:e2302580. [PMID: 37649226]
- Mon KKZ, Si Z, Chan-Park MB, and Kenney LJ. Polyimidazolium Protects against an Invasive Clinical Isolate of Salmonella Typhimurium. Antimicrob Agents Chemother 2022;:e0059722. [PMID: 36094258]
- Jacob H, Geng H, Shetty D, Halow N, Kenney LJ, and Nakano MM. Distinct Interaction Mechanism of RNAP and ResD and Distal Subsites for Transcription Activation of Nitrite Reductase in Bacillus subtilisψ. J Bacteriol 2021;:JB0043221. [PMID: 34898263]
- Singh MK, Zangoui P, Yamanaka Y, and Kenney LJ. Genetic code expansion enables visualization of Salmonella type three secretion system components and secreted effectors. Elife 2021; 10. [PMID: 34061032]
- Kenney LJ, and Anand GS. EnvZ/OmpR Two-Component Signaling: An Archetype System That Can Function Noncanonically. EcoSal Plus 2020; 9(1). [PMID: 32003321]
- Desai SK, and Kenney LJ. Switching Lifestyles Is an in vivo Adaptive Strategy of Bacterial Pathogens. Front Cell Infect Microbiol 2019; 9:421. [PMID: 31921700]
- Desai SK, Padmanabhan A, Harshe S, Zaidel-Bar R, and Kenney LJ. Salmonella biofilms program innate immunity for persistence in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 2019;. [PMID: 31160462]
- Ottemann KM, and Kenney LJ. Editorial overview: Host-pathogen interactions: bacteria. Curr. Opin. Microbiol. 2019; 47:iii-v. [PMID: 31138403]
Lab Members
CURRENT LAB MEMBERS
Liu Shuhan
Intern, Kenney Group
Xu Lisheng
PhD Student, Class of August 2016, Toyama Group, Kenney Group

About Linda J Kenney
The Kenney Lab pursues research in signal transduction and the regulation of gene expression in prokaryotes. In particular, they are studying the two-component regulatory system EnvZ/OmpR that regulates the expression of outer membrane proteins as well as many other genes. Their present work focuses on how OmpR activates genes required for systemic infection in Salmonella enterica.
Contact Us
Bacterial Pathogenesis & Signal Transduction Lab
Mechanobiology Institute (MBI)
National University of Singapore
T-Lab, #10-01
5A Engineering Drive 1
Singapore 117411
Phone: 6601 1285 ext 11285
Email: kenneyl@uic.edu
Web: mbi.nus.edu.sg/linda-j-kenney