Pulling biomolecular complexes
How pulling forces can counterintuitively stabilize proteins
Edited by Andrew MS Wong. Illustrations by Melanie Lee and Diego Pitta de Araujo | July 2019
A research team from the Mechanobiology Institute (MBI), National University of Singapore has revisited a decade-long outstanding question on the physical principles underlying a non-intuitive phenomenon, wherein pulling forces applied to some biomolecular complexes such as double-stranded DNA, or protein domains leads to their stabilization, rather than unravelling. The common consensus in the field is that such phenomenon can only be explained by complex multi-dimensional models. In contrast, the researchers in this study proved that force-induced stabilization of biomolecular complexes or protein domains can occur on a one-dimensional energy landscape, by taking into account extension fluctuations of the molecules during transition. The study was published in Communications Chemistry.

Artistic illustration of a double-stranded DNA biomolecular complex unfolding with increasing pulling force, depicted by arrows.
Understanding the mechanical lifetime of biomolecular complexes
Imagine a hook. Whether it stays firmly attached to its target depends on how much tension the hook is under. While it easily disconnects in the absence of stretching forces, it holds firmly as it is stretched more and more, until the stretching forces exceed a certain limit. This basic mechanical behaviour could be used to understand the non-intuitive effects of pulling forces on the mechanical lifetime of biomolecular complexes or protein domains.
Intuitively, pulling forces applied to protein domains or biomolecular complexes should decrease their lifetime, resulting in faster unfolding or separation of the domains that make up the complex (See schematic). This is because domain unfolding or complex separation is associated with extension elongation along the force direction, which decreases the potential energy. Such force-induced destabilization is termed “slip-bond” behaviour, and has been supported by results from studies that involve stretching of protein domains or biomolecular complexes using atomic force microscopy. Due to the rapid thermal and mechanical drift of most atomic force microscopy instruments, the measurements were typically captured over short experimental time scales by applying high forces, which typically do not exist in live cells, to accelerate the process of protein domain unfolding or complex separation.
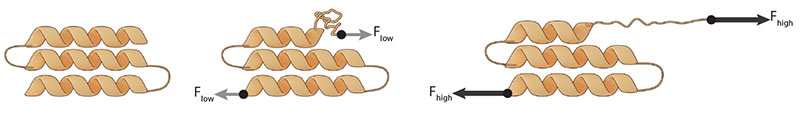
Protein domain unfolding or separation occurs with increasing force, along the force direction. This ‘slip-bond’ behaviour differs from ‘catch-bond’ behaviour, where increasing force leads to domain stabilization.
However, recent advent in single-molecule technologies have made it possible to conduct such experiments over physiologically relevant longer time scales. This has enabled researchers to measure the rate of domain unfolding or domains separation at a much lower, physiologically relevant, force range. Intriguingly, the lifetime of some protein domains or biomolecular complexes are known to increase at such low force range, where the higher the force applied, the longer it takes to unfold the protein domain or separate the complex. Such force-induced stabilization of biomolecular complexes or protein domains is referred to as “catch-bond” behaviour. Catch-bond behaviour is typically observed only over a certain force range – increasing the force beyond this range often causes a switch from catch-bond to slip-bond behaviour.
A new theoretical understanding explains how pulling forces can stabilize, rather than unfold, biomolecular complexes.
Understanding the physical principles that underlie catch-bond behaviour and the catch-to-slip switch has posed a challenge to theoreticians for decades. Previous studies have shown that these phenomena can be understood by assuming that force over a certain range may induce additional attractive interactions, which could stabilize the biomolecules and cause them to unfold or separate at a slower transition rate. Theoretically, these models imply that the transition energy landscape is multi-dimensional. While they can explain the experimental observations, it still remains unclear why catch-bond and catch-to-slip switch behaviours are predominantly observed at low forces.
The inferences from this current paper provide a plausible answer to this question. At low forces, the conformations of biomolecules undergo thermal fluctuations, with their extension lengths depending on the applied force. However, at high forces, which causes the biomolecules to be nearly fully extended, such force-dependent extension fluctuations can be neglected. In two earlier studies carried out in the lab (Yuan et al., Angew. Chem. 2017, 129, 5582; Guo et al., Chem. Sci. 2018, 9, 5871-5882), researchers had shown that such low-force extension fluctuations may result in increased energy barriers for some biomolecules under certain stretching conditions, leading to catch-bond and catch-to-slip switch behaviours. However, those studies did not address one key question: whether such behaviour can be understood on a one-dimensional energy landscape.
Explaining force-dependent stabilization
Tackling this specific question here, the researchers hypothesize that the catch-bond and catch-to-slip switch behaviours (taking into account the force-dependent extension fluctuations at low forces) could be explained by a simple one-dimensional model, with a properly defined transition coordinate. They suggest that the molecular extension change, which has been used as a transition coordinate for describing force-dependent energy landscapes in several previous studies, may be an inaccurate parameter at low forces due to the force-dependent extension fluctuations. They further show how properly chosen transition coordinates can lead to understanding catch-to-slip behaviour on a one-dimensional energy landscape, in a wide range of biomolecular systems. Thus, this work provides a novel perspective on the force-dependent stabilization of protein domains and biomolecular complexes.










