Building heart connections
How heart cells find their partners
Written by Sruthi Jagannathan | Illustration by Melanie Lee, PhD | June 2018
Latest research shows that filopodial activity regulated by distinctive distribution of cell adhesion molecules in heart cells is responsible for robust cell matching during Drosophila heart development. This study, led by MBI Principal Investigator Assistant Professor Timothy Saunders and graduate student Shaobo Zhang, was published in Developmental Cell.

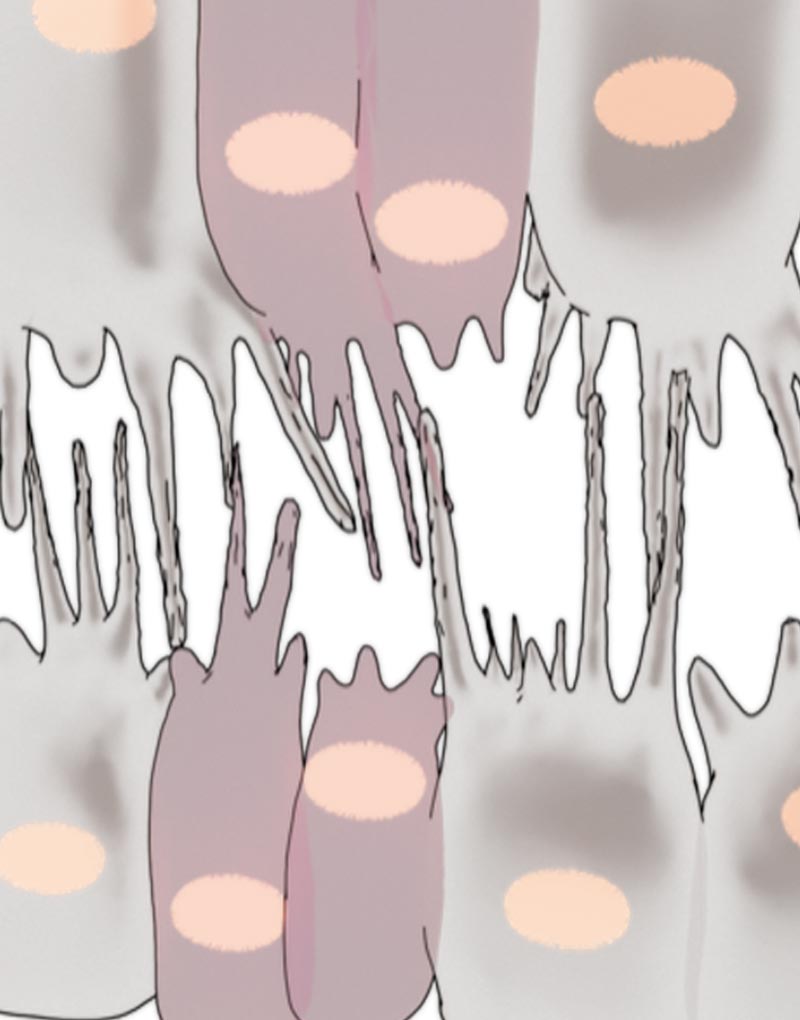
The schematic shows how cardioblasts use filopodia to probe cardioblasts on the opposing row in order to make physical connections with the right partner during Drosophila heart formation.
Cardioblasts use a robust cell recognition system to connect with partners
Congenital heart diseases are deformities that are present in the structure of the heart at birth. Characterized by malformations in heart walls or valves, or in the blood vessels carrying blood to or from the heart, they cause a wide range of disease conditions in newborns and young children that are often debilitating or even life-threatening. In order to come up with improved treatment or prevention strategies for these diseases, it is first important to understand how the shape of the heart develops and what factors may be important in the cases when development does not proceed normally.
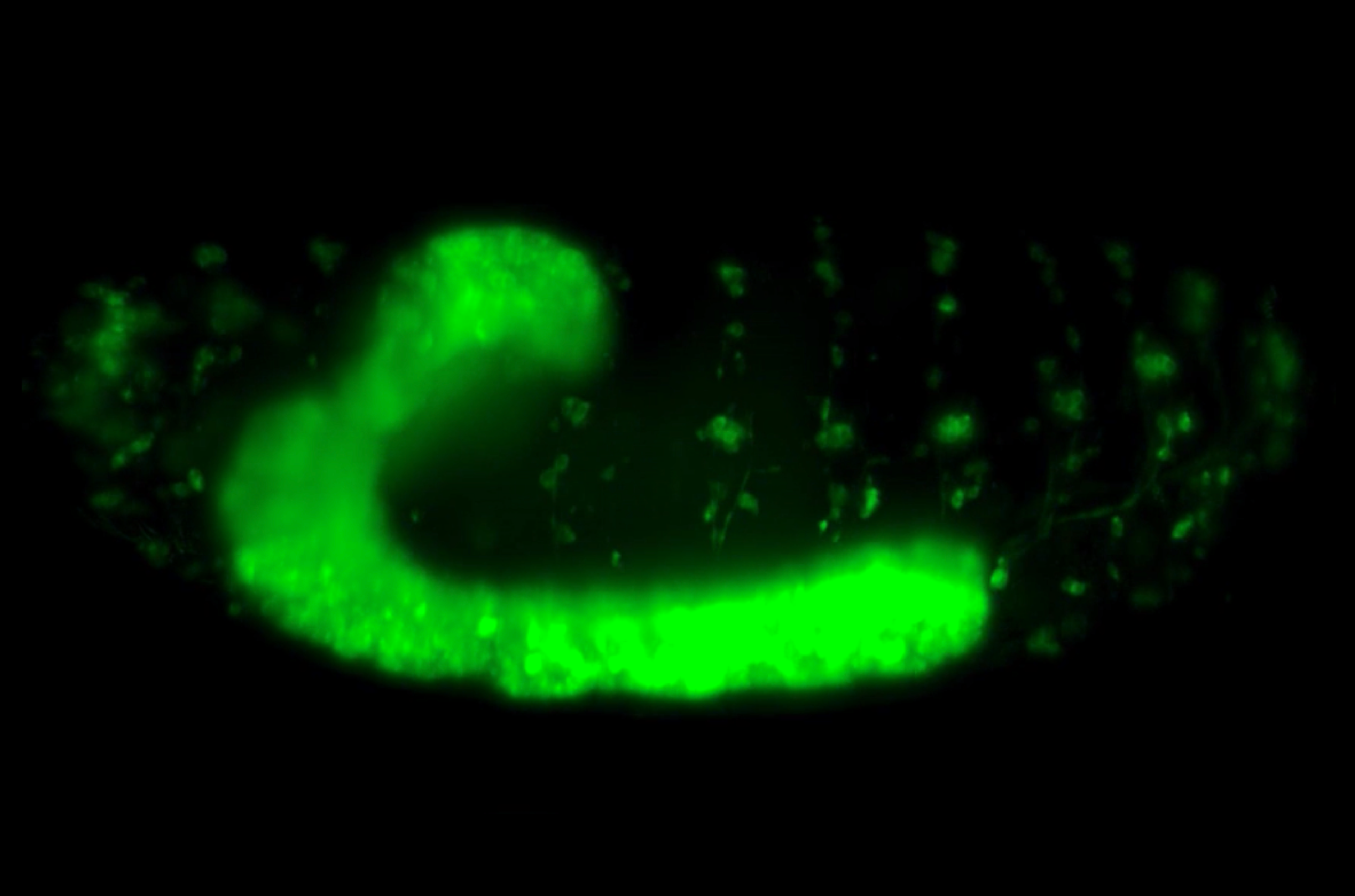
Heart development, like the development of other major organs in our body, is driven by the formation of precise physical connections between cells. During the very early stages of heart formation, two opposing rows of cardioblasts (heart cells) move towards each other, align side by side, and establish physical connections with partner cardioblasts on the opposing row. The buttoning up of the two rows of cardioblasts gives rise to primitive heart structures. This basic process occurs in many species, from fruit flies to humans.
By using confocal microscopy techniques to visualize live the process of heart formation in Drosophila (fruit fly) embryos, Zhang and team revealed how filopodia and CAMs work in concert to ensure a robust cell recognition system during Drosophila heart formation.
So how do cardioblasts find and connect with their right partners? This process is reliant on a robust cell recognition and matching system, where cardioblasts use ‘antennae’ to probe cardioblasts on the opposing row and form stable physical connections with the ones that they recognize as correct partners. These antennae are cellular structures called filopodia that form as spiky protrusions on cardioblast surfaces and carry specialized ‘molecular glue’ at their tips. The molecular glue, referred to as cell adhesion molecules (CAMs), help cells adhere to neighboring cells. But cell-to-cell adherence via CAMs is a highly selective process; as the filopodial tips probe their surroundings, CAMs look out for identical CAMs on the filopodial tips of neighboring cardioblasts. Once they encounter cardioblasts of their ‘type’ on the opposing row, they initiate physical connections between the filopodial tips. The connections then become stronger and more stable through ensuing cellular processes. When this cell recognition system fails, cardioblasts often connect to the wrong partner, and when several such mismatches line up in a row, they lead to a defective heart structure.
What makes the cell recognition system so robust? This question formed the basis of a recent study led by graduate student Shaobo Zhang from the lab of Assistant Professor Timothy Saunders at the Mechanobiology Institute, National University of Singapore. By using confocal microscopy techniques to visualize live the process of heart formation in Drosophila (fruit fly) embryos, Zhang and team attempted to find out how filopodia and CAMs work in concert to ensure a robust cell recognition system during Drosophila heart formation.
Of the several proteins that were screened as potential CAMs involved in heart formation, the researchers narrowed down their study to the functions of fascilin III (Fas3) and teneurin-m (Ten-m). Both these molecules displayed one important property: they were distributed non-uniformly in cardioblasts. The Drosophila heart is formed of two cardioblast subtypes; while Fas3 is present in high levels in one subtype, the other subtype contains high levels of Ten-m.
Depending on which CAM was present in high levels, the cardioblasts showed differences in how strongly they connected to their partners. While the filopodial contacts formed between two Fas3-rich cardioblasts were very strong and stable, those formed between Ten-m rich cardioblasts were relatively weak.
With these findings, the researchers were able to validate their hypothesis that filopodial activity is regulated by CAM distribution. They proposed important implications for the distinctive distribution pattern of CAMS: firstly, it forms the basis for a robust cell recognition system, wherein cardioblasts can recognize and form connections with correct partners on opposing rows; secondly, by influencing filopodial binding strengths, it also defines the course of cellular events that take place during Drosophila heart formation. Fas3-rich cardioblasts, which adhere strongly, lead the heart formation process, with the weakly binding Ten-m cardioblasts taking longer to stabilize.
This study describes how filopodia and CAMs work hand-in-hand to function as a robust cell recognition and matching system during heart development. Given the prevalence of congenital defects worldwide, the findings from this study will be valuable in predicting what goes wrong at the cellular and molecular level during incidences of heart malformations. Furthermore, the cellular machinery described here is not exclusive to heart development (filopodia and similar CAMs are present on various other cell types), and therefore could be extended to understand the inner workings of complex physiological systems such as the nervous system, where the lack of precise neural connections can lead to debilitating disorders like Parkinson’s, Alzheimer’s, and schizophrenia.