How is nuclear “prestress” maintained?
As an integral part of cellular behavior, cells are sensitive to matrix rigidity, local geometry and stress or strain applied by external factors [1]. In recent years, it has been established that an extensive network of protein assembly couples the cytoskeleton to the nucleus [2] (reviewed in [3]) and that condensation forces of the chromatin balance cytoskeletal forces resulting in a prestressed nuclear organization [4][5]. Hence, besides remodeling cytoskeletal filaments, the forces generated within the cell and that experienced at distant cell surface sites converge to the nucleus. This can happen either by physical transmission along the linked cytoskeleton [6] (reviewed in [3]) or by chemical signaling, where transcription regulators get transported to the nucleus upon activation [7][8] (reviewed in [9]). These mechanosignals have a significant impact on the mechanical properties of the nucleus such as shape and rigidity (reviewed in [6] [10]) through modification of the scaffolding proteins at the nuclear envelope and interior.
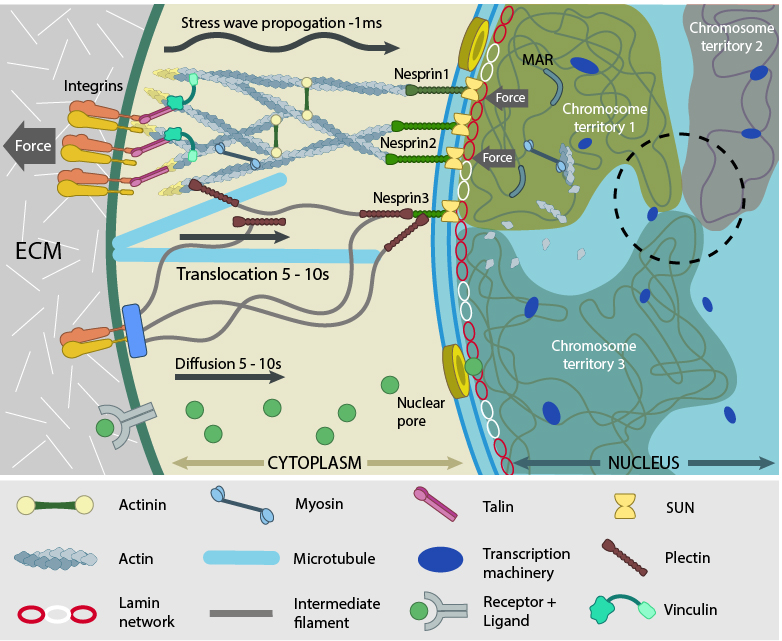
Force experienced by integrins at the cell surface via mechanosensing structures like focal adhesions (integrin cluster linked to actin network), hemidesmosomes (blue rectangle) or cell-cell contact (not shown) is accumulated, channeled through SUN1/SUN2 form the LINC (linker of nucleoskeleton and cytoskeleton) complexes connecting further to the nuclear lamina (red and white lamin network) and hence the attached nuclear scaffold proteins (actin and myosin). Chromatin attaches directly to the lamina and to other scaffolding proteins through the matrix attachment regions (MARs). Upon sensing the force, the nuclear scaffold help repositioning the chromatin thus affecting nuclear prestress and activating genes within milliseconds. Spatial segregation of chromosomes with defined territories is represented as colored compartments inside the nucleus. The dotted circle highlights looping of genes from different chromosomes to form a cluster in 3D space and share transcription apparatus (navy ovals). On the contrary, chemical signaling mediated by motor-based translocation along cytoskeletal filaments or diffusion of activated regulatory factors takes few seconds. Adapted from [19197334, 21391812]
With the cytoskeleton providing an outward pull and chromatin compaction providing an inward force, the nucleus can be envisioned as an elastic structure maintained in a stretched state due to force balance. Any perturbation to this homeostasis causes disorder in the system due to shrinkage or bulging of the nucleus, thus impairing its functional landscape. DNA processing and gene expression programs are affected due to alterations in cytoskeleton, nucleoskeleton and genome compaction [17] [4] (reviewed in [9]). In other words, the nucleus can be regarded as a load bearing organelle that can physically transmit mechanical cues [18][19] and a plethora of cellular traits, such as shape, motility, differentiation and development.
References
- Vogel V, and Sheetz M. Local force and geometry sensing regulate cell functions. Nat. Rev. Mol. Cell Biol. 2006; 7(4):265-75. [PMID: 16607289]
- Maniotis AJ, Chen CS, and Ingber DE. Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proc. Natl. Acad. Sci. U.S.A. 1997; 94(3):849-54. [PMID: 9023345]
- Stewart CL, Roux KJ, and Burke B. Blurring the boundary: the nuclear envelope extends its reach. Science 2007; 318(5855):1408-12. [PMID: 18048680]
- Mazumder A, Roopa T, Basu A, Mahadevan L, and Shivashankar GV. Dynamics of chromatin decondensation reveals the structural integrity of a mechanically prestressed nucleus. Biophys. J. 2008; 95(6):3028-35. [PMID: 18556763]
- Mazumder A, and Shivashankar GV. Emergence of a prestressed eukaryotic nucleus during cellular differentiation and development. J R Soc Interface 2010; 7 Suppl 3:S321-30. [PMID: 20356876]
- Wang N, Tytell JD, and Ingber DE. Mechanotransduction at a distance: mechanically coupling the extracellular matrix with the nucleus. Nat. Rev. Mol. Cell Biol. 2009; 10(1):75-82. [PMID: 19197334]
- Xu J, Zutter MM, Santoro SA, and Clark RA. A three-dimensional collagen lattice activates NF-kappaB in human fibroblasts: role in integrin alpha2 gene expression and tissue remodeling. J. Cell Biol. 1998; 140(3):709-19. [PMID: 9456329]
- Vartiainen MK, Guettler S, Larijani B, and Treisman R. Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactor MAL. Science 2007; 316(5832):1749-52. [PMID: 17588931]
- Shivashankar GV. Mechanosignaling to the cell nucleus and gene regulation. Annu Rev Biophys 2011; 40:361-78. [PMID: 21391812]
- Dahl KN, Ribeiro AJS, and Lammerding J. Nuclear shape, mechanics, and mechanotransduction. Circ. Res. 2008; 102(11):1307-18. [PMID: 18535268]
- Nagele RG, Freeman T, McMorrow L, Thomson Z, Kitson-Wind K, and Lee HY. Chromosomes exhibit preferential positioning in nuclei of quiescent human cells. J. Cell. Sci. 1999; 112 ( Pt 4):525-35. [PMID: 9914164]
- Marshall WF. Order and disorder in the nucleus. Curr. Biol. 2002; 12(5):R185-92. [PMID: 11882311]
- Webster M, Witkin KL, and Cohen-Fix O. Sizing up the nucleus: nuclear shape, size and nuclear-envelope assembly. J. Cell. Sci. 2009; 122(Pt 10):1477-86. [PMID: 19420234]
- Jorgensen P, Edgington NP, Schneider BL, Rupes I, Tyers M, and Futcher B. The size of the nucleus increases as yeast cells grow. Mol. Biol. Cell 2007; 18(9):3523-32. [PMID: 17596521]
- Wu C, Rolfe PA, Gifford DK, and Fink GR. Control of transcription by cell size. PLoS Biol. 2010; 8(11):e1000523. [PMID: 21072241]
- Versaevel M, Grevesse T, and Gabriele S. Spatial coordination between cell and nuclear shape within micropatterned endothelial cells. Nat Commun 2012; 3:671. [PMID: 22334074]
- Mazumder A, and Shivashankar GV. Gold-nanoparticle-assisted laser perturbation of chromatin assembly reveals unusual aspects of nuclear architecture within living cells. Biophys. J. 2007; 93(6):2209-16. [PMID: 17496030]
- Chiquet M, Gelman L, Lutz R, and Maier S. From mechanotransduction to extracellular matrix gene expression in fibroblasts. Biochim. Biophys. Acta 2009; 1793(5):911-20. [PMID: 19339214]
- Wang JH, Thampatty BP, Lin J, and Im H. Mechanoregulation of gene expression in fibroblasts. Gene 2007; 391(1-2):1-15. [PMID: 17331678]


