How is DNA replicated?
Cells must replicate their DNA before they can divide. This ensures that each daughter cell gets a copy of the genome, and therefore, successful inheritance of genetic traits. DNA replication is an essential process and the basic mechanism is conserved in all organisms. DNA replicates in the S phase of the cell cycle and initiates at specific regions in the DNA sequence known as DNA replication ‘origins’. A number of proteins participate in DNA replication and the process is subject to scrutiny by cell surveillance mechanisms called cell cycle checkpoints. These checkpoints ensure that replication of DNA occurs just once per cell cycle. Defects in DNA replication could give rise to damaging mutations including those that cause cancer.
The Initiation of DNA replication
The stage for DNA replication is set in the G1 phase of the cell cycle and DNA is synthesized in the S phase. DNA replication is initiated at specific sites in the genome known as the ‘origins’ which are recognized and bound by origin binding proteins. Replication commences at a single origin in prokaryotes and at multiple origins in eukaryotes, however, the basic mechanism of replication is conserved in all organisms [1]. In eukaryotes, initiator proteins ORC, Cdc6 and Cdt1 recruit the replicative helicase [2]. The eukaryotic replicative helicase is a complex of proteins called the CMG helicase consisting of Cdc45, Mcm2-7 and GINS proteins [3]. This assembly of the pre-replicative complex (pre-RC) at origins during G1 phase is called ‘origin licensing’ FIG. The helicase is inactive in the pre-RC and is activated only in the S phase when origins ‘fire’ due to the activity of CDK/DDK kinases [4], [5]. Once origins fire, DNA synthesis begins and the initiator proteins are degraded or exported out of the nucleus to prevent re-replication [6]. The precise mechanisms of origin licensing and origin firing in two separate phases of the cell-cycle ensure that DNA replication occurs only once per cell-cycle.
DNA synthesis
The mechanism of DNA replication is greatly influenced by DNA structure. The complementary base pairing between the nitrogen bases A-T and G-C underlies the semi-conservative nature of DNA replication, which results in a duplicated genome with one parental strand and one newly synthesized strand. Each strand serves as a template for the DNA polymerase to catalyze the addition of the correct base during synthesis of a new complementary strand. As the strands are antiparallel with opposing polarity and since DNA polymerases can only synthesize DNA in the 5′ to 3′ direction, only one strand is continuously synthesized. This strand is called the leading strand. Synthesis of the other strand, called the lagging strand, is made possible through discontinuous synthesis of short fragments, called Okazaki fragments, in the 5′ to 3′ direction, which are later joined together.
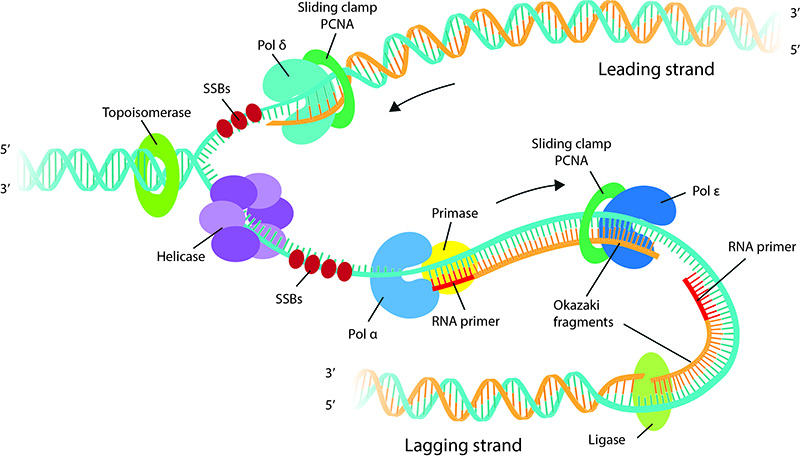
DNA replication: DNA replication proteins at the replication fork. The helicase unwinds the duplex DNA and Single Strand Binding proteins (SSBs) coat and stabilize single stranded DNA formed by strand separation. Topoisomerase is seen ahead of the fork removing superhelical tension caused by strand separation. Note that the leading strand is synthesized continuously in the 5′ to 3′ direction, whereas the lagging strand is synthesized discontinuously as short fragments called Okazaki fragments. The Polymerase α-primase complex synthesizes short RNA primers that are extended up to 30-40 nucleotides. Thereafter polymerase ε and polymerase δ takes up the job of faster and efficient strand synthesis on lagging and leading strands respectively. Ligase seals the gap between Okazaki fragments.
DNA synthesis begins in S phase as the replicative helicase unwinds and separates the two strands of the DNA double helix [7]. As the helicase unwinds DNA, DNA polymerase synthesizes DNA utilizing the exposed single stranded DNA as a template. DNA polymerases ‘read’ the template strand and add the correct complimentary base. Energy for polymerization comes from release of a pyrophosphate from a free deoxyribonucleotide triphosphate (dNTP), creating a 5′monophosphate that could be covalently linked to the 3′ hydroxyl group of another nucleotide. However, DNA polymerases cannot synthesize DNA de novo and require a preexisting primer with a free hydroxyl group to add nucleotides and extend the chain. A specialized RNA polymerase called primase synthesizes short RNA sequences about 10 nucleotides long which serve as primers. A single primer aids DNA replication on the leading strand and multiple primers initiate okazaki fragment synthesis on the lagging strand. In Eukaryotes, the primase is part of the DNA polymerase α (reviewed in [8]). The replicative helicase and primase functionally co-operate and stimulate each other’s activity [9].
After DNA polymerase α has synthesized a short, 30-40 nucleotide stretch of DNA, further DNA synthesis is handed over to polymerase ε and polymerase δ which have a higher processivity than polymerase α. The higher processivity or the ability of the polymerases to stay associated with DNA for upto 10kb without falling off is due to their association with a sliding clamp called PCNA. The polymerase switching enables DNA synthesis with high fidelity as polymerase ε and polymerase δ have a 3′ – 5′ exonuclease activity which enables proof reading and removal of any incorrect bases that is incorporated (reviewed in [8]). At the replication fork, there is a division of labor between the polymerases where polymerase ε carries out leading strand synthesis and polymerase δ is involved in the synthesis of the lagging strand [10], 12)
Okazaki fragment maturation and replication termination
The Okazaki fragments which are about 100-200bp in eukaryotes are ligated together in a process known as Okazaki fragment maturation to complete DNA synthesis. Polymerase δ, as it runs into the adjacent Okazaki fragment ahead of polymerization removes 2 to 3 nucleotides of the RNA primer thereby generating a short flap that is processed by Fen1 [11]. This leaves a nick that is sealed by DNA ligase1 [12]. Although there are well-defined replications termination sequences called Ter sites in prokaryotes, in eukaryotes, termination typically occurs by the collision of two replication forks.
DNA Replication: a mechanobiology perspective
DNA replication begins with the unwrapping and unwinding of the highly compacted chromatin structure. The two strands of the double helix must also be separated before the replication machinery can access and copy each strand. Specialized ATPase motor proteins called helicases catalyze DNA unwinding by translocating along the DNA substrate and separating the base pairs [13].
As DNA unwinding and DNA synthesis progresses, the DNA ahead of the replication fork becomes overwound or positively supercoiled. This creates superhelical tension which is usually resolved by enzymes known as topoisomerases. However, the super helical tension is higher in longer chromosomes [14] and in regions of the chromatin tethered to the nuclear envelope (reviewed in [15]). It is now evident that the torsional stress from the replication forks impinge on the nuclear envelope in the form of mechanical signals that recruit ATR, a DNA damage checkpoint protein, independent of its role in DNA repair [16]. ATR may then enable transient detachment of chromatin from the nuclear envelope, thus allowing for the completion of replication [16]. ATR is also recruited during prophase to resolve the topological stress arising from chromatin condensation and is required for coordinating DNA replication and chromatin condensation.
Aside from mechanical forces generated within the cell as a result of DNA replication itself, DNA replication may also be affected by external forces acting on the cell. It is well-known that external forces transduce to the nucleus via cytoskeletal links and affects gene regulation as well as organization of chromosomes [17]. Therefore understanding DNA replication requires a mechanobiology perspective, incorporating the physical challenges of DNA packaging and unwinding as well as mechanical forces that influence DNA replication.
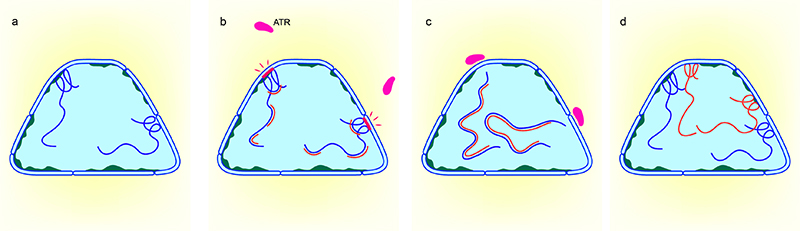
Model showing ATR-mediated checkpoint in response to mechanical stress created by DNA replication at the nuclear envelope: a) Nucleus showing DNA (dark blue strands) with regions tethered to the nuclear envelope b) Mechanical stress (red bar) created at the nuclear envelope by replicating DNA (red strands) c) Recruitment of ATR to the nuclear envelope transiently detaches DNA from the nuclear envelope allowing for completion of DNA replication. d) Nucleus showing newly replicated DNA (Adapted from Kumar et al, ATR mediates a checkpoint at the nuclear envelope in response to mechanical stress, Cell, 2014)
References
- O’Donnell M, Langston L, and Stillman B. Principles and concepts of DNA replication in bacteria, archaea, and eukarya. Cold Spring Harb Perspect Biol 2013; 5(7). [PMID: 23818497]
- Remus D, Beuron F, Tolun G, Griffith JD, Morris EP, and Diffley JFX. Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell 2009; 139(4):719-30. [PMID: 19896182]
- Langston LD, Zhang D, Yurieva O, Georgescu RE, Finkelstein J, Yao NY, Indiani C, and O’Donnell ME. CMG helicase and DNA polymerase ε form a functional 15-subunit holoenzyme for eukaryotic leading-strand DNA replication. Proc. Natl. Acad. Sci. U.S.A. 2014; 111(43):15390-5. [PMID: 25313033]
- Sheu Y, and Stillman B. The Dbf4-Cdc7 kinase promotes S phase by alleviating an inhibitory activity in Mcm4. Nature 2010; 463(7277):113-7. [PMID: 20054399]
- Zou L, and Stillman B. Formation of a preinitiation complex by S-phase cyclin CDK-dependent loading of Cdc45p onto chromatin. Science 1998; 280(5363):593-6. [PMID: 9554851]
- Hook SS, Lin JJ, and Dutta A. Mechanisms to control rereplication and implications for cancer. Curr. Opin. Cell Biol. 2007; 19(6):663-71. [PMID: 18053699]
- Vijayraghavan S, and Schwacha A. The eukaryotic Mcm2-7 replicative helicase. Subcell. Biochem. 2012; 62:113-34. [PMID: 22918583]
- Stillman B. DNA polymerases at the replication fork in eukaryotes. Mol. Cell 2008; 30(3):259-60. [PMID: 18471969]
- You Z, De Falco M, Kamada K, Pisani FM, and Masai H. The mini-chromosome maintenance (Mcm) complexes interact with DNA polymerase α-primase and stimulate its ability to synthesize RNA primers. PLoS ONE 2013; 8(8):e72408. [PMID: 23977294]
- Plosky BS. More division of labor at the eukaryotic replication fork. Mol. Cell 2014; 56(4):467-8. [PMID: 25459878]
- Balakrishnan L, and Bambara RA. Flap endonuclease 1. Annu. Rev. Biochem. 2013; 82:119-38. [PMID: 23451868]
- Balakrishnan L, and Bambara RA. Okazaki fragment metabolism. Cold Spring Harb Perspect Biol 2013; 5(2). [PMID: 23378587]
- Singleton MR, Dillingham MS, and Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 2007; 76:23-50. [PMID: 17506634]
- Kegel A, Betts-Lindroos H, Kanno T, Jeppsson K, Ström L, Katou Y, Itoh T, Shirahige K, and Sjögren C. Chromosome length influences replication-induced topological stress. Nature 2011; 471(7338):392-6. [PMID: 21368764]
- Bermejo R, Kumar A, and Foiani M. Preserving the genome by regulating chromatin association with the nuclear envelope. Trends Cell Biol. 2012; 22(9):465-73. [PMID: 22771046]
- Kumar A, Mazzanti M, Mistrik M, Kosar M, Beznoussenko GV, Mironov AA, Garrè M, Parazzoli D, Shivashankar GV, Scita G, Bartek J, and Foiani M. ATR mediates a checkpoint at the nuclear envelope in response to mechanical stress. Cell 2014; 158(3):633-46. [PMID: 25083873]
- Shivashankar GV. Mechanosignaling to the cell nucleus and gene regulation. Annu Rev Biophys 2011; 40:361-78. [PMID: 21391812]


