How do chromosome territory dynamics affect gene redistribution?
The spatial organization of chromatin within the 3-dimensional space of a chromosome territory enables the co-localization of co-transcribed genes and their transcriptional foci. Many gene positioning studies have shown that individual genes often loop out of their chromosomal territory to co-localize with transcription factories. This often leads to interchromosomal compartments becoming enriched with intermingling chromatin loops, either from the same chromosome, or different chromosomes. It has been suggested that this repositioning can occur upon transcriptional activation [1][2][3].
Measuring the diffusional motion of chromatin by sub-micrometer single-particle tracking, a characteristic confinement radius (significantly smaller than the size of the nucleus) can be determined for each locus. This demonstrates that, at least in yeast, centromers and telomers have a radii of confinement approximately twice as small as the rest of the chromosomal sites [4]. The authors also showed that in yeasts and drosophila, chromatin constantly undergoes diffusive Brownian motion, constrained by confinement regions of gene loci, which rarely exceed 0.3 µm [4].
Importantly, the repositioning of genes through chromosome territory dynamics is not always random, and the spatial redistribution of genes may involve specific nuclear structures or landmarks. This may have a significant effect on gene expression [5]. Local compaction dynamics, long-distance interactions with alternative sections of DNA, and interactions with nuclear scaffolds [6] all play a role in the control of gene redistribution. Where interactions between DNA and nuclear scaffolds occur, anchor points , known as matrix attachment regions (MARs), are formed.
Long-distance chromatin interactions may either involve the establishment of physical contacts between two sequence elements that are not adjacent to each other, but are present on the same linear chromosome (as is the case when enhancers interact with promoters), or between loci on different chromosomes [6][7]. Importantly, with most interactions between loci occurring in only a small fraction of cells at any given time, long-range contacts are considered to be, at least in part, random, and are therefore difficult to predict [8]. Despite these difficulties, it has been suggested that the nucleoskeleton is involved in the regulation of long-distance contacts. For example, Chuang et al. [9] observed a fast (0.1-0.9 µm/min) long-range (1-5 µm) directional movement of transgenic chromatin arrays. Nuclear actin together with myosin – two important components of the nucleoskeleton – were proposed to serve as molecular motors that direct the movement of chromatin towards a given target region [10]. This was supported when the movement of actin arrays was blocked by the expression of mutant nuclear myosin I or mutant actin. Moreover, with the actin mutant unable to polymerize, the looping of U2 snRNA genes towards coiled bodies was also abolished [11].
Interactions between genes and nuclear landmarks also affect gene transcription. These landmarks, which are distinct nuclear regions, include the nuclear lamina (NL), nuclear pore complexes (NPCs) and the nucleolus (reviewed in [12][13][14]).
The nuclear periphery is often found to preferentially interact with transcriptionally silent chromatin, which is characterized by a low gene density. It has been proposed that the nuclear periphery itself creates a specific environment that favors histone deacetylation and gene silencing [15]. Indeed, in yeast cells (which generally lack nuclear lamin), gene silencing resulted from the tethering of a gene locus to the periphery [15]. This suggests that in mammals, the nuclear periphery is heterogeneous, with microdomains of different compositions having different effects on genome function [12]. Takizawa et al. [5] proposed that simply being near the periphery without physically associating with the NL is not enough to induce gene repression.
Nuclear pore complexes represent another distinct microenvironment; however, in contrast to the nuclear periphery this landmark is often associated with gene activation [16][17]. In yeast cells, the highly transcribed ribosomal protein (RP) is connected with NPCs via the actin-related protein, Arp6 [18]. Interestingly, interactions between chromatin and NPCs may take place both in and away from the nuclear periphery, making the dynamic movement of lamins and nuclear pore proteins integral to gene regulation [19]. The exact dynamics that drive these interactions in the nucleoplasm remains poorly understood.
The nucleolus may also anchor specific chromatin loci. In addition to rRNA genes, it often harbors large genomic regions (median size 750 kb) that are enriched in centromeric satellite repeats and inactive gene clusters. Centromeric regions are also found associated with nuclear lamina, suggesting that centromeres are distributed between the nuclear lamina and nucleoli [14].
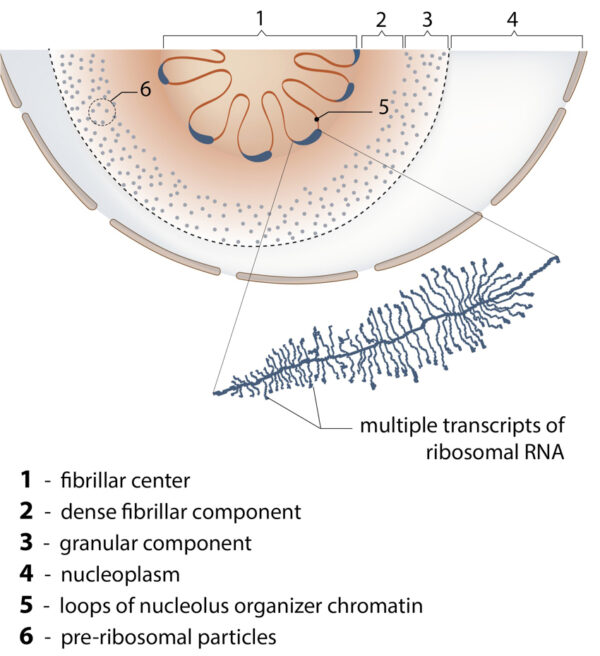
The main function of nucleolus is the synthesis of ribosomal RNA and assembly of ribosomal particles. Nucleoli are often enriched with centromeric satellite repeats and inactive gene clusters.
Despite the current evidence highlighting the influence of chromosome territory dynamics in the regulation of gene expression, little is known on the mechanisms behind these processes. For example, it is unclear how and why activated genes are translocated, or loop, from one chromosome territory to another. One plausible scenario involves the transduction of mechanical stimuli to the nucleus directly via the cytoskeleton. In such cases, cytoskeletal forces induce nuclear deformation (i.e., elongation or squeeze) and subsequently, alter chromosome topology and gene expression [20]. The nucleoskeleton (i.e., nuclear actin and myosin) provides another mechanism to control the long-distance directional movement of genes [10], but this is likely to be restricted to specific genes, e.g. U2 snRNA [11]. Currently, the extent to which genes move through active guidance as opposed to diffusion, remains unclear.
References
- Volpi EV, Chevret E, Jones T, Vatcheva R, Williamson J, Beck S, Campbell RD, Goldsworthy M, Powis SH, Ragoussis J, Trowsdale J, and Sheer D. Large-scale chromatin organization of the major histocompatibility complex and other regions of human chromosome 6 and its response to interferon in interphase nuclei. J. Cell. Sci. 2000; 113 ( Pt 9):1565-76. [PMID: 10751148]
- Wiblin AE, Cui W, Clark AJ, and Bickmore WA. Distinctive nuclear organisation of centromeres and regions involved in pluripotency in human embryonic stem cells. J. Cell. Sci. 2005; 118(Pt 17):3861-8. [PMID: 16105879]
- Osborne CS, Chakalova L, Mitchell JA, Horton A, Wood AL, Bolland DJ, Corcoran AE, and Fraser P. Myc dynamically and preferentially relocates to a transcription factory occupied by Igh. PLoS Biol. 2007; 5(8):e192. [PMID: 17622196]
- Marshall WF, Straight A, Marko JF, Swedlow J, Dernburg A, Belmont A, Murray AW, Agard DA, and Sedat JW. Interphase chromosomes undergo constrained diffusional motion in living cells. Curr. Biol. 1997; 7(12):930-9. [PMID: 9382846]
- Takizawa T, Meaburn KJ, and Misteli T. The meaning of gene positioning. Cell 2008; 135(1):9-13. [PMID: 18854147] van Steensel B. Chromatin: constructing the big picture. EMBO J. 2011; 30(10):1885-95. [PMID: 21527910]
- Hakim O, Sung M, and Hager GL. 3D shortcuts to gene regulation. Curr. Opin. Cell Biol. 2010; 22(3):305-13. [PMID: 20466532]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO,
- Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES, and Dekker J. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 2009; 326(5950):289-93. [PMID: 19815776]
- Chuang C, Carpenter AE, Fuchsova B, Johnson T, de Lanerolle P, and Belmont AS. Long-range directional movement of an interphase chromosome site. Curr. Biol. 2006; 16(8):825-31. [PMID: 16631592]
- Fedorova E, and Zink D. Nuclear architecture and gene regulation. Biochim. Biophys. Acta 2008; 1783(11):2174-84. [PMID: 18718493]
- Dundr M, Ospina JK, Sung M, John S, Upender M, Ried T, Hager GL, and Matera AG. Actin-dependent intranuclear repositioning of an active gene locus in vivo. J. Cell Biol. 2007; 179(6):1095-103. [PMID: 18070915]
- Deniaud E, and Bickmore WA. Transcription and the nuclear periphery: edge of darkness? Curr. Opin. Genet. Dev. 2009; 19(2):187-91. [PMID: 19231154]
- Towbin BD, Meister P, and Gasser SM. The nuclear envelope–a scaffold for silencing? Curr. Opin. Genet. Dev. 2009; 19(2):180-6. [PMID: 19303765]
- van Steensel B, and Dekker J. Genomics tools for unraveling chromosome architecture. Nat. Biotechnol. 2010; 28(10):1089-95. [PMID: 20944601]
- Andrulis ED, Neiman AM, Zappulla DC, and Sternglanz R. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature 1998; 394(6693):592-5. [PMID: 9707122]
- Casolari JM, Brown CR, Komili S, West J, Hieronymus H, and Silver PA. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 2004; 117(4):427-39. [PMID: 15137937]
- Capelson M, Doucet C, and Hetzer MW. Nuclear pore complexes: guardians of the nuclear genome. Cold Spring Harb. Symp. Quant. Biol. 2011; 75:585-97. [PMID: 21502404]
- Yoshida T, Shimada K, Oma Y, Kalck V, Akimura K, Taddei A, Iwahashi H, Kugou K, Ohta K, Gasser SM, and Harata M. Actin-related protein Arp6 influences H2A.Z-dependent and -independent gene expression and links ribosomal protein genes to nuclear pores. PLoS Genet. 2010; 6(4):e1000910. [PMID: 20419146]
- Van Bortle K, and Corces VG. Spinning the web of cell fate. Cell 2013; 152(6):1213-7. [PMID: 23498930]
- Gieni RS, and Hendzel MJ. Mechanotransduction from the ECM to the genome: are the pieces now in place? J. Cell. Biochem. 2008; 104(6):1964-87. [PMID: 17546585]


